Abstract
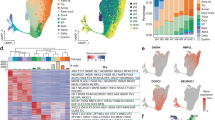
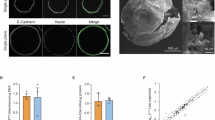
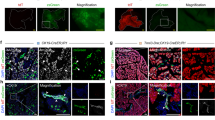
Cell fate decisions in human endoderm development are tightly regulated, yet the role of metabolic products remains elusive. The endodermal posterior foregut gives rise to pancreas, liver, and intestine. Here, we identify Glutathione Peroxidase 2 as a critical regulator of human posterior foregut differentiation, revealing oxidative stress as a key determinant of pancreatic versus non-pancreatic cell fate. Cells lacking Glutathione Peroxidase 2 under pancreas-promoting conditions differentiate also into hepatic-like progenitors. Through bulk and single-cell transcriptomics, chromatin accessibility profiling, and functional studies, we reveal that Glutathione Peroxidase 2 orchestrates lineage commitment by regulating key transcription factors, leading to emergence of multilineage liver and intestinal progenitors. Mechanistically, Glutathione Peroxidase 2 deficiency triggers extracellular matrix remodeling, activating bone morphogenetic protein signaling and skewing differentiation from the pancreatic lineage. Manipulating oxidative stress recapitulates or rescues Glutathione Peroxidase 2 loss effects, establishing oxidative stress as a gatekeeper of pancreatic fate. Controlling oxidative stress during in vitro differentiation could advance regenerative medicine applications.
Similar content being viewed by others
Data availability
All data generated or analyzed during this study are available. Processed data and all datasets necessary to interpret the results are included in the published article and its Supplementary Information. Raw RNA-seq, scRNA-seq, and ATAC-Seq data generated during the study have been deposited in the NCBI GEO database under accession number GSE291135. Source data are provided as a Source Data file. Raw image files are available from the corresponding author upon a reasonable request. The authors declare that cell lines are available for the research community upon request from the corresponding author. Other published datasets we used in this study could be obtained from: E-MTAB-696719, OMIX00161636, GSE197064107, GSM268939920, E-MTAB-8210 and E-MTAB-71899. Source data are provided with this paper.
Code availability
R code generated for RNA-seq and scRNA-seq data analysis are available at GitHub (https://github.com/WJSzlachcic/BorowiakLab_2025_GPX2/).
References
Zaret, K. S. & Grompe, M. Generation and regeneration of cells of the liver and pancreas. Science 322, 1490–1494 (2008).
Sambathkumar, R. et al. Generation of hepatocyte- and endocrine pancreatic-like cells from human induced endodermal progenitor cells. PLoS ONE 13, e0197046 (2018).
Yiangou, L., Ross, A. D. B., Goh, K. J. & Vallier, L. Human pluripotent stem cell-derived endoderm for modeling development and clinical applications. Cell Stem Cell 22, 485–499 (2018).
Willnow, D. et al. Quantitative lineage analysis identifies a hepato-pancreato-biliary progenitor niche. Nature 597, 87–91 (2021).
Koike, H. et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary. Nature 574, 112–116 (2019).
De Franco, E. et al. Primate-specific ZNF808 is essential for pancreatic development in humans. Nat. Genet. 55, 2075–2081 (2023).
Yang, D. et al. CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction. Nat. Cell Biol. 24, 1064–1076 (2022).
Loh, K. M. et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14, 237–252 (2014).
Wesley, B. T. et al. Single-cell atlas of human liver development reveals pathways directing hepatic cell fates. Nat. Cell Biol. 24, 1487–1498 (2022).
Leenders, F. et al. Oxidative stress leads to β-cell dysfunction through loss of β-cell identity. Front. Immunol. 12, 690379 (2021).
Sies, H. & Jones, D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21, 363–383 (2020).
Petrova, B. et al. Dynamic redox balance directs the oocyte-to-embryo transition via developmentally controlled reactive cysteine changes. Proc. Natl. Acad. Sci. USA 115, E7978–E7986 (2018).
Agca, C., Klein, W. H. & Venuti, J. M. Reduced O2 and elevated ROS in sea urchin embryos leads to defects in ectoderm differentiation. Dev. Dyn. 238, 1777–1787 (2009).
Coffman, J. A., McCarthy, J. J., Dickey-Sims, C. & Robertson, A. J. Oral-aboral axis specification in the sea urchin embryo II. Mitochondrial distribution and redox state contribute to establishing polarity in Strongylocentrotus purpuratus. Dev. Biol. 273, 160–171 (2004).
Amblard, I. et al. H2O2 and Engrailed 2 paracrine activity synergize to shape the zebrafish optic tectum. Commun. Biol. 3, 1–9 (2020).
Wang, Y.-C. et al. Non-enzymatic role of SOD1 in intestinal stem cell growth. Cell Death Dis. 13, 882 (2022).
Chmielowiec, J. et al. Human pancreatic microenvironment promotes β-cell differentiation via non-canonical WNT5A/JNK and BMP signaling. Nat. Commun. 13, 1952 (2022).
Ziojła, N. M. et al. ETVs dictate hPSC differentiation by tuning biophysical properties. Nat. Commun. 16, 1999 (2025).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Scavuzzo, M. A. et al. Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat. Commun. 9, 3356 (2018).
Wang, P., Rodriguez, R. T., Wang, J., Ghodasara, A. & Kim, S. K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell 8, 335–346 (2011).
Scheibner, K. et al. Epithelial cell plasticity drives endoderm formation during gastrulation. Nat. Cell Biol. 23, 692–703 (2021).
Viotti, M., Nowotschin, S. & Hadjantonakis, A.-K. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 16, 1146–1156 (2014).
Saad, R. S., Ghorab, Z., Khalifa, M. A. & Xu, M. CDX2 as a marker for intestinal differentiation: Its utility and limitations. World J. Gastrointest. Surg. 3, 159–166 (2011).
Raghoebir, L. et al. SOX2 redirects the developmental fate of the intestinal epithelium toward a premature gastric phenotype. J. Mol. Cell Biol. 4, 377–385 (2012).
DeLaForest, A. et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 138, 4143–4153 (2011).
Ito, R. et al. Elucidation of HHEX in pancreatic endoderm differentiation using a human iPSC differentiation model. Sci. Rep. 13, 8659 (2023).
Ebrahim, N., Shakirova, K. & Dashinimaev, E. PDX1 is the cornerstone of pancreatic β-cell functions and identity. Front. Mol. Biosci. 9, 1091757 (2022).
Dusing, M. R., Florence, E. A. & Wiginton, D. A. Pdx-1 is required for activation in vivo from a duodenum-specific enhancer. J. Biol. Chem. 276, 14434–14442 (2001).
Kawaguchi, Y. Sox9 and programming of liver and pancreatic progenitors. J. Clin. Investig. 123, 1881–1886 (2013).
Wong, Y. F. et al. Expansion of ventral foregut is linked to changes in the enhancer landscape for organ-specific differentiation. Nat. Cell Biol. 25, 481–492 (2023).
Ee, L. S. et al. Enhancer remodeling by OTX2 directs specification and patterning of mammalian definitive endoderm. Dev. Cell 60, 3431–3445.e8 (2025).
Wells, J. M. & Melton, D. A. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development 127, 1563–1572 (2000).
Andriopoulos, B. et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 41, 482–487 (2009).
Funa, N. S. et al. TGF-β modulates cell fate in human ES cell-derived foregut endoderm by inhibiting Wnt and BMP signaling. Stem Cell Rep. 19, 973–992 (2024).
Ma, Z. et al. Deciphering early human pancreas development at the single-cell level. Nat. Commun. 14, 5354 (2023).
Jiang, W. et al. CD24: a novel surface marker for PDX1-positive pancreatic progenitors derived from human embryonic stem cells. Stem Cells 29, 609–617 (2011).
Bushman, T. L. & Kuemmerle, J. F. IGFBP-3 and IGFBP-5 production by human intestinal muscle: reciprocal regulation by endogenous TGF-beta1. Am. J. Physiol. 275, G1282–1290 (1998).
Kuemmerle, J. F. & Zhou, H. Insulin-like growth factor-binding protein-5 (IGFBP-5) stimulates growth and IGF-I secretion in human intestinal smooth muscle by Ras-dependent activation of p38 MAP kinase and Erk1/2 pathways. J. Biol. Chem. 277, 20563–20571 (2002).
Flynn, R. S., Mahavadi, S., Murthy, K. S., Kellum, J. M. & Kuemmerle, J. F. Insulin-like growth factor-binding protein-5 stimulates growth of human intestinal muscle cells by activation of Gαi3. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G1232–G1238 (2009).
Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018).
Sekiya, M., Hiraishi, A., Touyama, M. & Sakamoto, K. Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochem. Biophys. Res. Commun. 375, 602–607 (2008).
Lee, J., Homma, T., Kurahashi, T., Kang, E. S. & Fujii, J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem. Biophys. Res. Commun. 464, 229–235 (2015).
Ehrhart, M., Grube, D., Bader, M. F., Aunis, D. & Gratzl, M. Chromogranin A in the pancreatic islet: cellular and subcellular distribution. J. Histochem. Cytochem. 34, 1673–1682 (1986).
Sander, M. et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 127, 5533–5540 (2000).
Luzi, L., Zerbini, G. & Caumo, A. C-peptide: a redundant relative of insulin? Diabetologia 50, 500–502 (2007).
Watt, A. J., Garrison, W. D. & Duncan, S. A. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 37, 1249–1253 (2003).
Ang, L. T. et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 22, 2190–2205 (2018).
Veres, A. et al. Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Suzuki, A., Sekiya, S., Büscher, D., Izpisúa Belmonte, J. C. & Taniguchi, H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development 135, 1589–1595 (2008).
Lüdtke, T. H.-W., Christoffels, V. M., Petry, M. & Kispert, A. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology 49, 969–978 (2009).
Zhang, Y., Feng, X. H. & Derynck, R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 394, 909–913 (1998).
Chen, W. et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Mukherjee, S., French, D. L. & Gadue, P. Loss of TBX3 enhances pancreatic progenitor generation from human pluripotent stem cells. Stem Cell Rep. 16, 2617–2627 (2021).
Montgomery, R. K. et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA 108, 179–184 (2011).
Middelhoff, M. et al. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am. J. Physiol. Gastrointest. Liver Physiol. 313, G285–G299 (2017).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011).
Hynes, R. O. & Naba, A. Overview of the matrisome-an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4, a004903 (2012).
Kopec, A. K. & Luyendyk, J. P. Coagulation in liver toxicity and disease: role of hepatocyte tissue factor. Thromb. Res. 133, S57–S59 (2014).
Muciño-Bermejo, J., Carrillo-Esper, R., Uribe, M. & Méndez-Sánchez, N. Coagulation abnormalities in the cirrhotic patient. Ann. Hepatol. 12, 713–724 (2013).
Berthier, A., Johanns, M., Zummo, F. P., Lefebvre, P. & Staels, B. PPARs in liver physiology. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166097 (2021).
Lisman, T. & Luyendyk, J. P. Platelets as modulators of liver diseases. Semin. Thromb. Hemost. 44, 114–125 (2018).
Asantewaa, G. et al. Glutathione synthesis in the mouse liver supports lipid abundance through NRF2 repression. Nat. Commun. 15, 6152 (2024).
Luo, J., Yang, H. & Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225–245 (2020).
Nemes, K., Åberg, F., Gylling, H. & Isoniemi, H. Cholesterol metabolism in cholestatic liver disease and liver transplantation: from molecular mechanisms to clinical implications. World J. Hepatol. 8, 924–932 (2016).
Gough, N. R., Xiang, X. & Mishra, L. TGF-β signaling in liver, pancreas, and gastrointestinal diseases and cancer. Gastroenterology 161, 434–452.e15 (2021).
Wu, M., Wu, S., Chen, W. & Li, Y.-P. The roles and regulatory mechanisms of TGF-β and BMP signaling in bone and cartilage development, homeostasis and disease. Cell Res. 34, 101–123 (2024).
Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2, vbac016 (2022).
Cheng, S. K., Olale, F., Brivanlou, A. H. & Schier, A. F. Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2, E30 (2004).
Chen, X. et al. BMP and activin receptor membrane bound inhibitor: BAMBI has multiple roles in gene expression and diseases (Review). Exp. Ther. Med. 27, 28 (2023).
Ramirez, F. & Sakai, L. Y. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 339, 71–82 (2010).
Tsutsui, K. et al. ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J. Biol. Chem. 285, 4870–4882 (2010).
Shi, Y. & Massagué, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003).
Macías-Silva, M. et al. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87, 1215–1224 (1996).
Kretzschmar, M., Liu, F., Hata, A., Doody, J. & Massagué, J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 11, 984–995 (1997).
Derynck, R. & Budi, E. H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal 12, eaav5183 (2019).
Lennicke, C. et al. Loss of epithelium-specific GPx2 results in aberrant cell fate decisions during intestinal differentiation. Oncotarget 9, 539–552 (2018).
Sherwood, R. I., Chen, T.-Y. A. & Melton, D. A. Transcriptional dynamics of endodermal organ formation. Dev. Dyn. 238, 29–42 (2009).
Di Marzo, N., Chisci, E. & Giovannoni, R. The role of hydrogen peroxide in redox-dependent signaling: homeostatic and pathological responses in mammalian cells. Cells 7, 156 (2018).
Hoarau, E., Chandra, V., Rustin, P., Scharfmann, R. & Duvillie, B. Pro-oxidant/antioxidant balance controls pancreatic β-cell differentiation through the ERK1/2 pathway. Cell Death Dis. 5, e1487 (2014).
Knutson, A. K., Williams, A. L., Boisvert, W. A. & Shohet, R. V. HIF in the heart: development, metabolism, ischemia, and atherosclerosis. J. Clin. Investig. 131, e137557 (2021).
Murata, H. et al. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J. Biol. Chem. 278, 50226–50233 (2003).
Sarbassov, D. D. & Sabatini, D. M. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 280, 39505–39509 (2005).
Yoshida, S. et al. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J. Biol. Chem. 286, 32651–32660 (2011).
Chaudhari, P., Ye, Z. & Jang, Y.-Y. Roles of reactive oxygen species in the fate of stem cells. Antioxid. Redox Signal 20, 1881–1890 (2014).
D’Oria, R. et al. The role of oxidative stress in cardiac disease: from physiological response to injury factor. Oxid. Med. Cell Longev. 2020, 5732956 (2020).
Manda-Handzlik, A. & Demkow, U. Neutrophils: the role of oxidative and nitrosative stress in health and disease. Adv. Exp. Med. Biol. 857, 51–60 (2015).
Nakamura, H. & Takada, K. Reactive oxygen species in cancer: current findings and future directions. Cancer Sci. 112, 3945–3952 (2021).
Morrisey, E. E. et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12, 3579–3590 (1998).
Watt, A. J., Zhao, R., Li, J. & Duncan, S. A. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev. Biol. 7, 37 (2007).
Allen, H. L. et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat. Genet. 44, 20–22 (2011).
Carrasco, M., Delgado, I., Soria, B., Martín, F. & Rojas, A. GATA4 and GATA6 control mouse pancreas organogenesis. J. Clin. Investig. 122, 3504–3515 (2012).
Shaw-Smith, C. et al. GATA4 mutations are a cause of neonatal and childhood-onset diabetes. Diabetes 63, 2888–2894 (2014).
Yamagata, K. et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384, 458–460 (1996).
Chen, W. S. et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8, 2466–2477 (1994).
Ma, X. et al. N6-methyladenosine modification-mediated mRNA metabolism is essential for human pancreatic lineage specification and islet organogenesis. Nat. Commun. 13, 4148 (2022).
Velazco-Cruz, L. et al. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Rep. 12, 351–365 (2019).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Hafemeister, C. & Halbritter, F. Single-cell RNA-seq differential expression tests within a sample should use pseudo-bulk data of pseudo-replicates. Preprint at https://doi.org/10.1101/2023.03.28.534443 (2023).
Borcherding, N. et al. Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun. Biol. 4, 122 (2021).
Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Müller-Dott, S. et al. Expanding the coverage of regulons from high-confidence prior knowledge for accurate estimation of transcription factor activities. Nucleic Acids Res. 51, 10934–10949 (2023).
Xie, Z. et al. Gene set knowledge discovery with EnrichR. Curr. Protoc. 1, e90 (2021).
Clarke, D. J. B. et al. Appyters: turning Jupyter notebooks into data-driven web apps. Patterns 2, 100213 (2021).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Olaniru, O. E. et al. Single-cell transcriptomic and spatial landscapes of the developing human pancreas. Cell Metab. 35, 184–199.e5 (2023).
Kim, S.-I. et al. Inducible transgene expression in human IPS cells using versatile all-in-one piggyBac transposons. Methods Mol. Biol. 1357, 111–131 (2016).
Yusa, K., Zhou, L., Li, M. A., Bradley, A. & Craig, N. L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA 108, 1531–1536 (2011).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Acknowledgements
We would like to express our gratitude to Dr. Artur Jankowski for his excellent technical support. We also thank Dr. Ludovic Vallier and all members of the Borowiak lab for valuable discussions, comments, and support. We also thank Dr. Allan Bradley and Dr. Knut Woltjen for sharing plasmids via Addegene. This work was supported by the Polish National Science Center grant OPUS (2020/37/B/NZ3/01917 and 2020/39/B/NZ3/01408 to M. B.), Foundation for Polish Science and EU TEAM Programme (POIR.04.04.00-00-20C5/16-00) to M. B., Polish National Science Center, Miniatura grant (2022/06/X/NZ3/00465) to J. Sz., Polish National Science Center, Sonata grant (2021/43/D/NZ3/02294) to W. J. Sz., and Sonata grant (2022/47/D/NZ3/02068) to K. B.
Author information
Authors and Affiliations
Contributions
J.S.—experimental design and execution, including GPX2 KO hPSC generation, directed and spontaneous differentiation, scRNA-seq analysis of SC-β cells WT and KO cells, data acquisition and analysis, figure preparation, and manuscript writing; W. J.S.—scRNA-seq analysis of PFG WT and KO cells, figure preparation, and manuscript writing; K.B.—bulk RNA-sequencing and initial analysis, ATAC-seq initial analysis; M.S.—performed and analyzed ATAC-seq; M.Baginska.—participated in experimental execution; M.Borowiak.—experimental design, data analysis, manuscript writing, and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Saiyong Zhu who co-reviewed with Xiaojie Ma; Xiaojie Ma and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szpotkowska, J., Szlachcic, W.J., Blaszczyk, K. et al. Oxidative stress and GPX2 control pancreatic vs. non-pancreatic cell fate in human endoderm. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68145-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68145-x