Abstract
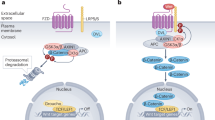
Protein phase separation has emerged as a crucial mechanism for spatiotemporal regulation of intracellular processes, yet its potential to integrate and compute diverse extracellular signals is not fully understood. Here, we show a mechano-biochemical circuit that harnesses phase separation to process mechanical and biochemical inputs, modulating cell fate decisions. We demonstrate that volumetric compression bidirectionally regulates canonical Wnt/β-catenin signaling, where the presence of Wnt ligands determines the locations of AXIN phase separation to form either LRP6 signalosomes on the cell membrane or β-catenin destruction complexes in the cytosol, while the mechanical stimulus promotes degree of phase separation to amplify either the positive or negative signal. This circuit enhances healthy intestinal organoid proliferation while suppressing patient-derived colorectal cancer organoid growth, revealing its potential for precise mechanotherapy. Our findings establish phase separation as a critical component in mechanical signal transduction and provide a framework for integrating mechanical and biochemical cues in cellular decision-making. This approach opens avenues for targeted therapies and deepens our understanding of how cells process complex environmental information.
Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences. Specifically, the single-cell RNA-seq data for human colorectal cancer organoids are accessible in GSA-Human under accession codes HRA015319 and HRA013862, and the bulk RNA-seq data are accessible under accession code HRA013863. The single-cell RNA-seq data for murine organoids are accessible under accession code CRA032833. All unique materials and stable cell lines generated in this study are available from the corresponding author upon reasonable request. All other data supporting the findings of this study are available within the article and its Supplementary Information. Source data are provided with this paper.
References
Li, Y., Tang, W. & Guo, M. The cell as matter: connecting molecular biology to cellular functions. Matter 4, 1863–1891 (2021).
Li, Y., Wong, I. Y. & Guo, M. Reciprocity of cell mechanics with extracellular stimuli: emerging opportunities for translational medicine. Small 18, 2107305 (2022).
Du, H. et al. Tuning immunity through tissue mechanotransduction. Nat. Rev. Immunol. 23, 174–188 (2023).
Romani, P., Valcarcel-Jimenez, L., Frezza, C. & Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 22, 22–38 (2021).
Saraswathibhatla, A., Indana, D. & Chaudhuri, O. Cell–extracellular matrix mechanotransduction in 3D. Nat. Rev. Mol. Cell Biol. 24, 495–516 (2023).
Di, X. et al. Cellular mechanotransduction in health and diseases: from molecular mechanism to therapeutic targets. Signal Transduct. Target. Ther. 8, 282 (2023).
Shyer, A. E. et al. Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science 357, 811–815 (2017).
Segel, M. et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 573, 130–134 (2019).
Li, Y. et al. Compression-induced dedifferentiation of adipocytes promotes tumor progression. Sci. Adv. 6, eaax5611 (2020).
Blache, U. et al. Engineered hydrogels for mechanobiology. Nat. Rev. Methods Prim. 2, 98 (2022).
Benham-Pyle, B. W., Pruitt, B. L. & Nelson, W. J. Mechanical strain induces E-cadherin–dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015).
Fernandez-Sanchez, M. E. et al. Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature 523, 92–95 (2015).
Li, Y. et al. Volumetric compression induces intracellular crowding to control intestinal organoid growth via Wnt/β-catenin signaling. Cell Stem Cell 28, 63–78.e67 (2021).
Jansen, J. H. et al. Stretch-induced inhibition of Wnt/β-catenin signaling in mineralizing osteoblasts. J. Orthop. Res. 28, 390–396 (2010).
Andreu, I. et al. Mechanical force application to the nucleus regulates nucleocytoplasmic transport. Nat. Cell Biol. 24, 896–905 (2022).
Esposito, D. et al. ROCK1 mechano-signaling dependency of human malignancies driven by TEAD/YAP activation. Nat. Commun. 13, 703 (2022).
Totaro, A. et al. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206 (2017).
Xue, X. et al. Mechanics-guided embryonic patterning of neuroectoderm tissue from human pluripotent stem cells. Nat. Mater. 17, 633–641 (2018).
Wang, N., Butler, J. P. & Ingber, D. E. Mechanotransduction across the cell surface and through the cytoskeleton. Science 260, 1124–1127 (1993).
Moroni, M., Servin-Vences, M. R., Fleischer, R., Sánchez-Carranza, O. & Lewin, G. R. Voltage gating of mechanosensitive PIEZO channels. Nat. Commun. 9, 1096 (2018).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Li, P. et al. High-throughput and proteome-wide discovery of endogenous biomolecular condensates. Nat. Chem. 16, 1101–1112 (2024).
MacDonald, B. T., Tamai, K. & He, X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009).
Müller, I. E. et al. Gene networks that compensate for crosstalk with crosstalk. Nat. Commun. 10, 4028 (2019).
Ahrends, R. et al. Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science 344, 1384–1389 (2014).
Zhu, R., del Rio-Salgado, J. M., Garcia-Ojalvo, J. & Elowitz, M. B. Synthetic multistability in mammalian cells. Science 375, eabg9765 (2022).
Xie, M. & Fussenegger, M. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 19, 507–525 (2018).
Bahrami-Nejad, Z. et al. A transcriptional circuit filters oscillating circadian hormonal inputs to regulate fat cell differentiation. Cell Metab. 27, 854–868.e858 (2018).
Bonnet, J., Yin, P., Ortiz, M. E., Subsoontorn, P. & Endy, D. Amplifying genetic logic gates. Science 340, 599–603 (2013).
Zhou, Z. et al. Engineering longevity—design of a synthetic gene oscillator to slow cellular aging. Science 380, 376–381 (2023).
Bienz, M. Head-to-tail polymerization in the assembly of biomolecular condensates. Cell 182, 799–811 (2020).
Nong, J. et al. Phase separation of Axin organizes the β-catenin destruction complex. J. Cell Biol. 220, e202012112 (2021).
Wang, Y. et al. USP10 strikes down β-catenin by dual-wielding deubiquitinase activity and phase separation potential. Cell Chem. Biol. 30, 1436–1452.e1410 (2023).
Fiedler, M., Mendoza-Topaz, C., Rutherford, T. J., Mieszczanek, J. & Bienz, M. Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating β-catenin. Proc. Natl. Acad. Sci. USA 108, 1937–1942 (2011).
Schwarz-Romond, T. et al. The DIX domain of dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 14, 484–492 (2007).
Wang, S. et al. Small-molecule modulation of Wnt signaling via modulating the Axin-LRP5/6 interaction. Nat. Chem. Biol. 9, 579–585 (2013).
Stamos, J. L. & Weis, W. I. The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898 (2013).
Bilic, J. et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 (2007).
Zeng, X. et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438, 873–877 (2005).
Kim, S.-E. et al. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867–870 (2013).
Li, V. S. et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 149, 1245–1256 (2012).
Hernández, A. R., Klein, A. M. & Kirschner, M. W. Kinetic responses of β-catenin specify the sites of Wnt control. Science 338, 1337–1340 (2012).
Liu, C. et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 (2002).
Morin, P. J. et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275, 1787–1790 (1997).
Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 (2014).
Law, S. M. & Zheng, J. J. Premise and peril of Wnt signaling activation through GSK-3β inhibition. Iscience 25, 104159 (2022).
Ring, D. B. et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52, 588–595 (2003).
Batlle, E. et al. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111, 251–263 (2002).
De Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Valenta, T. et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918 (2016).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
Jorissen, R. N. et al. Wild-type APC predicts poor prognosis in microsatellite-stable proximal colon cancer. Br. J. Cancer 113, 979–988 (2015).
Garcia-Reyes, B. et al. Discovery of inhibitor of Wnt production 2 (IWP-2) and related compounds as selective ATP-competitive inhibitors of casein kinase 1 (CK1) δ/ε. J. Med. Chem. 61, 4087–4102 (2018).
Ichiki, T. et al. Sensory representation and detection mechanisms of gut osmolality change. Nature 602, 468–474 (2022).
Li, Y. & Guo, M. Volumetric compression for engineering living systems. Nat. Rev. Bioeng. 2, 1023–1038 (2024).
Ng, K. M. et al. Single-strain behavior predicts responses to environmental pH and osmolality in the gut microbiota. MBio 14, e00753–00723 (2023).
Tropini, C. et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 173, 1742–1754.e1717 (2018).
Zimmerman, C. A. et al. A gut-to-brain signal of fluid osmolarity controls thirst satiation. Nature 568, 98–102 (2019).
Acknowledgements
We gratefully acknowledge the financial support from the National Key Research and Development Program of China (2024YFF1207300 to Y.L.), the National Natural Science Foundation of China (Grant numbers 32171248 to Y.L., 12472319 to Y.L.), the Fundamental Research Funds for Central Universities, HUST (2021GCRC056 to Y.L.).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.L.; Methodology: Y.L., J.S., L.W., P.L., F.Q., M.L., L.X., C.S., Y. Zheng, and Y. Zhang; Investigation: J.S., Y.L., X.R., H.X., and W.W.; Visualization: J.S., Y.L., and P.C.; Funding acquisition: Y.L.; Project administration: Y.L.; Supervision: Y.L. and B.F.L.; Writing—original draft: Y.L. and J.S.; Writing—review & editing: Y.L., J.S., and B.F.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Danfeng Cai and Shige Yoshimura for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, J., Wu, L., Li, P. et al. Volumetric compression regulates the phase separation of AXIN and acts as an operational amplifier to bidirectionally modulate Wnt signaling in organoids. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68209-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68209-y