Abstract
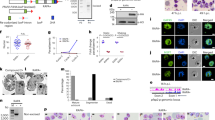
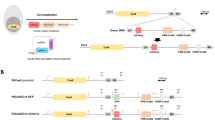
Malaria is caused by the proliferation of Plasmodium parasites in the vertebrate host blood stream through repeated cycles of asexual multiplication inside erythrocytes. During these cycles, parasites dynamically change their transcriptome at each developmental step to express genes exactly when required; however, the mechanisms regulating these transcriptomic changes remain unclear. In this study, we reveal that the AP2-family transcription factor PbAP2-TR is essential for the asexual blood stage development of the rodent malaria parasite Plasmodium berghei, as a transcriptional repressor. Conditional knockout of pbap2-tr causes developmental arrest at the trophozoite stage, i.e., the cell growth phase of asexual blood stage development. The expression of PbAP2-TR target genes is upregulated in pbap2-tr-knockout parasites, and introduction of mutations into the binding motifs increases the promoter activity of the target genes. Time-course transcriptome analysis shows that PbAP2-TR is largely responsible for the transcriptional downregulation from early to late trophozoite development. Furthermore, our data demonstrate that PbAP2-TR recruits a putative chromatin remodeler, PbMORC, as a co-factor. These results indicate that transcriptional repression by PbAP2-TR-PbMORC complex contributes on precise control of transcription peak patterns during the asexual blood stage development.
Similar content being viewed by others
Data availability
All ChIP-sequencing, RNA-sequencing and DIP-sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession numbers GSE290541, GSE290542, GSE290544, and GSE290545. All MS/MS data for RIME have been deposited in the ProteomeXchange Consortium under dataset PXD067426. Source data are provided with this paper.
References
World Health Organization. World malaria report 2024. https://www.who.int/publications/i/item/9789240104440 (2024).
Bannister, L. H. & Mitchell, G. H. The malaria merozoite, forty years on. Parasitology 136, 1435–1444 (2009).
Venugopal, K., Hentzschel, F., Valkiūnas, G. & Marti, M. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat. Rev. Microbiol. 2020 18, 177–189 (2020).
Voß, Y., Klaus, S., Guizetti, J. & Ganter, M. Plasmodium schizogony, a chronology of the parasite’s cell cycle in the blood stage. PLoS Pathog. 19, e1011157 (2023).
Bannister, L. H., Hopkins, J. M., Fowler, R. E., Krishna, S. & Mitchell, G. H. A brief illustrated guide to the ultrastructure of Plasmodium falciparum asexual blood stages. Parasitol. Today 16, 427–433 (2000).
LANGRETH, S. G., JENSEN, J. B., REESE, R. T. & TRAGER, W. Fine Structure of Human Malaria In Vitro. J. Protozool. 25, 443–452 (1978).
Marti, M., Good, R. T., Rug, M., Knuepfer, E. & Cowman, A. F. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science (1979) 306, 1930–1933 (2004).
Olszewski, K. L. et al. Host-parasite interactions revealed by plasmodium falciparum metabolomics. Cell Host Microbe 5, 191–199 (2009).
Rinehart, M. T., Park, H. S., Walzer, K. A., Chi, J. T. A. & Wax, A. Hemoglobin consumption by P. falciparum in individual erythrocytes imaged via quantitative phase spectroscopy. Sci. Rep. 2016 6, 1–9 (2016).
Lu, X. M. et al. Nascent RNA sequencing reveals mechanisms of gene regulation in the human malaria parasite Plasmodium falciparum. Nucleic Acids Res 45, 7825–7840 (2017).
Arnot, D. E., Ronander, E. & Bengtsson, D. C. The progression of the intra-erythrocytic cell cycle of Plasmodium falciparum and the role of the centriolar plaques in asynchronous mitotic division during schizogony. Int J. Parasitol. 41, 71–80 (2011).
Klaus, S. et al. Asynchronous nuclear cycles in multinucleated Plasmodium falciparum facilitate rapid proliferation. Sci. Adv. 8, 5362 (2022).
Rudlaff, R. M., Kraemer, S., Marshman, J. & Dvorin, J. D. Three-dimensional ultrastructure of Plasmodium falciparum throughout cytokinesis. PLoS Pathog. 16, e1008587 (2020).
Bozdech, Z. et al. The transcriptome of the intraerythrocytic developmental cycle of plasmodium falciparum. PLoS Biol. 1, e5 (2003).
Otto, T. D. et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Med. 12, 1–18 (2014).
Otto, T. D. et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol 76, 12–24 (2010).
Toenhake, C. G. et al. Chromatin accessibility-based characterization of the gene regulatory network underlying plasmodium falciparum blood-stage development. Cell Host Microbe 23, 557–569.e9 (2018).
Watzlowik, M. T. et al. Plasmodium blood stage development requires the chromatin remodeller Snf2L. Nature 2025 639, 1069–1075 (2025). 639:8056.
Balaji, S., Madan Babu, M., Iyer, L. M. & Aravind, L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006 (2005).
Singhal, R., Prata, I. O., Bonnell, V. A. & Llinás, M. Unraveling the complexities of ApiAP2 regulation in Plasmodium falciparum. Trends Parasitol. 40, 987–999 (2024).
Santos, J. M. et al. Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor. Cell Host Microbe 21, 731–741.e10 (2017).
Nishi, T., Kaneko, I. & Yuda, M. SIP2 is the master transcription factor of Plasmodium merozoite formation. Sci. Adv. 11, 5458 (2025).
Yuda, M., Sakaida, H. & Chinzei, Y. Targeted disruption of the Plasmodium berghei CTRP gene reveals its essential role in malaria infection of the vector mosquito. J. Exp. Med. 190, 1711–1715 (1999).
Modrzynska, K. et al. A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe 21, 11–22 (2017).
Bushell, E. et al. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170, 260–272.e8 (2017).
Zhang, C. et al. Systematic CRISPR-Cas9-mediated modifications of plasmodium yoelii ApiAP2 genes reveal functional insights into parasite development. mBio 8, 1–17 (2017).
Shinzawa, N. et al. Improvement of CRISPR/Cas9 system by transfecting Cas9-expressing Plasmodium berghei with linear donor template. Commun. Biol. 2020 3, 1–13 (2020).
Jullien, N., Sampieri, F., Enjalbert, A. & Herman, J. P. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res 31, e131–e131 (2003).
Knuepfer, E., Napiorkowska, M., Van Ooij, C. & Holder, A. A. Generating conditional gene knockouts in Plasmodium – a toolkit to produce stable DiCre recombinase-expressing parasite lines using CRISPR/Cas9. Sci. Rep. 2017 7, 1–12 (2017).
Shang, X. et al. A cascade of transcriptional repression determines sexual commitment and development in Plasmodium falciparum. Nucleic Acids Res 49, 9264–9279 (2021).
Campbell, T. L., de Silva, E. K., Olszewski, K. L., Elemento, O. & Llinás, M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 6, e1001165 (2010).
Alvarez-Jarreta, J. et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center in 2023. Nucleic Acids Res 52, D808–D816 (2024).
Painter, H. J. et al. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 2018 9, 1–12 (2018).
Singh, M. K. et al. A Plasmodium falciparum MORC protein complex modulates epigenetic control of gene expression through interaction with heterochromatin. Elife 12, RP92201 (2024).
Chahine, Z. M. et al. PfMORC protein regulates chromatin accessibility and transcriptional repression in the human malaria parasite, Plasmodium falciparum. Elife 12, RP92499 (2024).
Subudhi, A. K. et al. DNA-binding protein PfAP2-P regulates parasite pathogenesis during malaria parasite blood stages. Nat. Microbiol. 2023 8, 2154–2169 (2023).
Yuda, M., Iwanaga, S., Kaneko, I. & Kato, T. Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc. Natl. Acad. Sci. USA 112, 12824–12829 (2015).
Singh, S. et al. The PfAP2-G2 transcription factor is a critical regulator of gametocyte maturation. Mol. Microbiol 115, 1005–1024 (2021).
Nishi, T., Kaneko, I., Iwanaga, S. & Yuda, M. PbAP2-FG2 and PbAP2R-2 function together as a transcriptional repressor complex essential for Plasmodium female development. PLoS Pathog. 19, e1010890 (2023).
Sierra-Miranda, M. et al. PfAP2Tel, harbouring a non-canonical DNA-binding AP2 domain, binds to Plasmodium falciparum telomeres. Cell Microbiol 19, e12742 (2017).
Martins, R. M. et al. An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci. Rep. 2017 7, 1–10 (2017).
Cubillos, E. F. G., Prata, I. O., Fotoran, W. L., Ranford-Cartwright, L. & Wunderlich, G. The Transcription Factor PfAP2-O Influences Virulence Gene Transcription and Sexual Development in Plasmodium falciparum. Front Cell Infect. Microbiol 11, 669088 (2021).
Guizetti, J. & Scherf, A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol 15, 718–726 (2013).
Fraschka, S. A. et al. Comparative heterochromatin profiling reveals conserved and unique epigenome signatures linked to adaptation and development of malaria parasites. Cell Host Microbe 23, 407–420.e8 (2018).
Gomes, A. R. et al. A transcriptional switch controls sex determination in Plasmodium falciparum. Nature 2022 612, 528–533 (2022). 7940 612.
Schmoller, K. M. & Skotheim, J. M. The biosynthetic basis of cell size control. Trends Cell Biol. 25, 793–802 (2015).
Gilson, P. R., Chisholm, S. A., Crabb, B. S. & de Koning-Ward, T. F. Host cell remodelling in malaria parasites: a new pool of potential drug targets. Int J. Parasitol. 47, 119–127 (2017).
Counihan, N. A., Modak, J. K. & de Koning-Ward, T. F. How malaria parasites acquire nutrients from their host. Front Cell Dev. Biol. 9, 649184 (2021).
Wichers, J. S. et al. Dissecting the gene expression, localization, membrane topology, and function of the plasmodium falciparum STEVOR protein family. mBio 10, e01500-19 (2019).
Kono, M. et al. Pellicle formation in the malaria parasite. J. Cell Sci. 129, 673–680 (2016).
Yahata, K. et al. Gliding motility of Plasmodium merozoites. Proc. Natl. Acad. Sci. USA 118, e2114442118 (2021).
He, L. et al. Plasmodium falciparum GAP40 plays an essential role in merozoite invasion and gametocytogenesis. Microbiol Spectr. 11, e0143423 (2023).
Koch, A. et al. MORC proteins: Novel players in plant and animal health. Front Plant Sci. 8, 1720 (2017).
Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol 5, 570 (2020).
Fan, F., Xue, L., Yin, X., Gupta, N. & Shen, B. AP2XII-1 is a negative regulator of merogony and presexual commitment in Toxoplasma gondii. mBio 14, e0178523 (2023).
Antunes, A. V. et al. In vitro production of cat-restricted Toxoplasma pre-sexual stages. Nature 2023 625, 366–376 (2023). 7994 625.
Wang, J. L. et al. The transcription factor AP2XI-2 is a key negative regulator of Toxoplasma gondii merogony. Nat. Commun. 2024 15, 1–17 (2024).
Xue, L. et al. A novel nuclear protein complex controlling the expression of developmentally regulated genes in Toxoplasma Gondii. Adv. Sci. 12, 2412000 (2025).
Sinha, A. et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257 (2014).
Kafsack, B. F. C. et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 (2014).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Yuda, M. et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol 71, 1402–1414 (2009).
Yuda, M., Iwanaga, S., Shigenobu, S., Kato, T. & Kaneko, I. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol 75, 854–863 (2010).
Yuda, M., Kaneko, I., Murata, Y., Iwanaga, S. & Nishi, T. Mechanisms of triggering malaria gametocytogenesis by AP2-G. Parasitol. Int 84, 102403 (2021).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Iwanaga, S. et al. Functional identification of the plasmodium centromere and generation of a plasmodium artificial chromosome. Cell Host Microbe 7, 245–255 (2010).
Acknowledgements
This work was supported by the Japan Agency for Medical Research and Development (253fa627002h to S.I.), the Japan Society for the Promotion of Science (24K10187 to TN; 23H02709 to YM; 23K06515 to IK), and the Grant for Joint Research Project of the Research Institute for Microbial Diseases, The University of Osaka (JRPRIMD25B9 to IK). We would like to express our gratitude to Dr. Hiroko Kato and Dr. Ryohei Narumi, Central Instrumentation Laboratory, Research Institute for Microbial Diseases, The University of Osaka for performing analysis using timsTOF Pro and providing insight and expertise that greatly assisted the research.
Author information
Authors and Affiliations
Contributions
T.N. and Y.M. designed the study. T.N. performed all experiments, except for next generation sequencing and liquid chromatography-tandem mass spectrometry analysis. These analyses were performed by DNAFORM and Central Instrumentation Laboratory, Research Institute for Microbial Diseases, The University of Osaka, respectively. T.N. performed all formal data analyses and statistical analyses. I.K. contributed to the generation of transgenic parasites. S.I. contributed technical assistance/suggestions. All obtained funding for the study. Y.M. supervised the project. T.N. and Y.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mohamed Hakim, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nishi, T., Kaneko, I., Iwanaga, S. et al. Precise gene regulation through transcriptional repression is essential for Plasmodium berghei asexual blood stage development. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68222-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68222-1