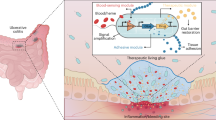
Abstract
The intestinal epithelium plays a critical role in health and disease, yet the impact of microenvironmental cues in diseased contexts, such as inflammatory bowel disease (IBD), remains poorly defined. To address this gap, we first benchmarked human colonic organoid injury models against IBD tissue and established a disease-relevant model of inflammation using inflammatory cytokines. Using this system, we built a dictionary of epithelial responses to 79 secreted niche factors at single cell resolution via donor-pooled, multiplexed single cell RNA-sequencing. The comprehensive nature of our atlas allowed us to map relationships between perturbations, infer the function of less characterized ligands, and identify cell type-specific perturbed pathways. Finally, we established the relevance of organoid-derived gene programs by mapping them to single cell and spatial atlases of human IBD tissue. Our resource offers a global view of epithelial responses to microenvironmental cues, offering insights into epithelial homeostasis and repair mechanisms in IBD.
Similar content being viewed by others
Data availability
The single-cell RNA-sequencing raw and processed data generated for this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the GEO Series accession number GSE313368. Source Data are provided with this paper. Data from the Parikh et al. study is deposited in the GEO database under accession number GSE116222. Data from the Kong et al. study are available for download from the controlled-access data repository, Broad DUOS (Accession DUOS-000146 CD_Atlas_2021_GIDER; DUOS-000145 CD_Atlas_2021_PRISM). Data from the Smillie et al. study are available at Single Cell Portal under accession number SCP259. Data from the Elmentaite et al. study is available at ArrayExpress with accession numbers E-MTAB-9543, E-MTAB-9536, E-MTAB-9532, E-MTAB-9533 and E-MTAB-10386. Data from the Yu et al. study is available at ArrayExpress with accession numbers E-MTAB-10187 and E-MTAB-10268. Source data are provided with this paper.
Code availability
Code related to this manuscript is available at github.com/Genentech/secretome_dictionary.
References
Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013).
Abud, H. E., Amarasinghe, S. L., Micati, D. & Jardé, T. Stromal niche signals that orchestrate intestinal regeneration. Cell. Mol. Gastroenterol. Hepatol. 17, 679–685 (2024).
Abraham, C. & Cho, J. H. Inflammatory bowel disease. N. Engl. J. Med. 361, 2066–2078 (2009).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386.e17 (2018).
Thomas, T. et al. A longitudinal single-cell atlas of anti-tumour necrosis factor treatment in inflammatory bowel disease. Nat. Immunol. 25, 2152–2165 (2024).
Holloway, E. M. et al. Mapping development of the human intestinal niche at single-cell resolution. Cell Stem Cell 28, 568–580.e4 (2021).
Wen, C., Chen, D., Zhong, R. & Peng, X. Animal models of inflammatory bowel disease: category and evaluation indexes. Gastroenterol. Rep. 12, goae021 (2024).
Kiesler, P., Fuss, I. J. & Strober, W. Experimental Models of Inflammatory Bowel Diseases. CMGH 1, 154–170 (2015).
Ferreira, B. et al. Trends in 3D models of inflammatory bowel disease. Biochim. Biophys. Acta Mol. Basis Dis. 1870, 167042 (2024).
Meir, M. et al. Enteroids generated from patients with severe inflammation in Crohn’s disease maintain alterations of junctional proteins. J. Crohns Colitis. 14, 1473–1487 (2020).
Arnauts, K. et al. Ex vivo mimicking of inflammation in organoids derived from patients with ulcerative colitis. Gastroenterology 159, 1564–1567 (2020).
Beumer, J. et al. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022).
Chen, L. et al. TGFB1 induces fetal reprogramming and enhances intestinal regeneration. Cell Stem Cell 30, 1520–1537.e8 (2023).
Lukonin, I. et al. Phenotypic landscape of intestinal organoid regeneration. Nature 586, 275–280 (2020).
Zhang, J. et al. Tahoe-100M: a giga-scale single-cell perturbation atlas for context-dependent gene function and cellular modeling. Preprint at https://www.biorxiv.org/content/10.1101/2025.02.20.639398v1 (2025).
Sanchís-Calleja, V. O. P. et al. Decoding morphogen patterning of human neural organoids with a multiplexed single-cell transcriptomic screen. Preprint at https://www.biorxiv.org/content/10.1101/2024.02.08.579413v1 (2024).
Cui, A. et al. Dictionary of immune responses to cytokines at single-cell resolution. Nature 625, 377–384 (2023).
Ayyaz, A. et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 596, 121–125 (2019).
Orzechowska-Licari, E. J., LaComb, J. F., Giarrizzo, M., Yang, V. W. & Bialkowska, A. B. Intestinal epithelial regeneration in response to ionizing irradiation. J. Vis. Exp. 185, 64028 (2023)
Montenegro-Miranda, P. S. et al. A novel organoid model of damage and repair identifies HNF4α as a critical regulator of intestinal epithelial regeneration. Cell. Mol. Gastroenterol. Hepatol. 10, 209–223 (2020).
Katsandegwaza, B., Horsnell, W. & Smith, K. Inflammatory bowel disease: a review of pre-clinical murine models of human disease. Int. J. Mol. Sci. 23, 9344 (2022).
Macedo, M. H., Neto, M. D., Pastrana, L., Gonçalves, C. & Xavier, M. Recent advances in cell-based in vitro models to recreate human intestinal inflammation. Adv. Sci. 10, 2301391 (2023).
Aherne, C. M. et al. Neuronal guidance molecule netrin-1 attenuates inflammatory cell trafficking during acute experimental colitis. Gut 61, 695–705 (2012).
Kong, L. et al. The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity 56, 444–458.e5 (2023).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
Yu, Q. et al. Charting human development using a multi-endodermal organ atlas and organoid models. Cell 184, 3281–3298.e22 (2021).
Brandenberg, N. et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 863–874 (2020)
Wang, Q. et al. Cytokine-induced epithelial permeability changes are regulated by the activation of the p38 mitogen-activated protein kinase pathway in cultured Caco-2 cells. Shock 29, 531–537 (2008).
Antoni, L., Nuding, S., Wehkamp, J. & Stange, E. F. Intestinal barrier in inflammatory bowel disease. World J. Gastroenterol. 20, 1165–1179 (2014).
Iwamoto, M., Koji, A., Makiyama, K., Kobayashi, N. & Nakane, P. K. Apoptosis of crypt epithelial cells in ulcerative colitis. J. Pathol. 180, 152–159 (1996).
Michielan, A. & D’Incà, R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediat. Inflamm. 2015, 628157 (2015)
d’Aldebert, E. et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front. Cell Dev. Biol. 8, 363 (2020).
Co, J. Y. et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520.e4 (2019).
Oost, K. C. et al. Dynamics and plasticity of stem cells in the regenerating human colonic epithelium. Preprint at https://www.biorxiv.org/content/10.1101/2023.12.18.572103v1 (2023).
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y. & Wrana, J. L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015).
Yui, S. et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49.e7 (2018).
Nusse, Y. M. et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113 (2018).
Li, J. et al. Identification and multimodal characterization of a specialized epithelial cell type associated with Crohn’s disease. Nat. Commun. 15, 7204 (2024).
Grasberger, H. et al. DUOX2 variants associate with preclinical disturbances in microbiota-immune homeostasis and increased inflammatory bowel disease risk. J. Clin. Invest. 131, e141676 (2021).
Bolton, C. et al. An integrated taxonomy for monogenic inflammatory bowel disease. Gastroenterology 162, 859–876 (2022).
Song, F. et al. The role of alcohol dehydrogenase 1C in regulating inflammatory responses in ulcerative colitis. Biochem. Pharm. 192, 114691 (2021).
Santhanam, S. et al. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm. Bowel Dis. 18, 2158–2168 (2012).
MacFie, T. S. et al. DUOX2 and DUOXA2 form the predominant enzyme system capable of producing the reactive oxygen species H2O2 in active ulcerative colitis and are modulated by 5-aminosalicylic acid. Inflamm. Bowel Dis. 20, 514–524 (2014).
Vatn, S. S. et al. Mucosal gene transcript signatures in treatment naïve inflammatory bowel disease: a comparative analysis of disease to symptomatic and healthy controls in the European IBD-character cohort. Clin. Exp. Gastroenterol. 15, 5–25 (2022).
Schniers, A. et al. Ulcerative colitis: functional analysis of the in-depth proteome. Clin Proteomics 16, 4 (2019).
Barnich, N. et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574 (2007).
Parikh, K. et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55 (2019).
Abud, H. E., Chan, W. H. & Jardé, T. Source and Impact of the EGF Family of Ligands on Intestinal Stem Cells. Front. Cell Dev. Biol. 9, 685665 (2021).
Keir, M., Yi, T., Lu, T. & Ghilardi, N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 217, e20192195 (2020).
Wagner, F. et al. Dose escalation randomised study of efmarodocokin alfa in healthy volunteers and patients with ulcerative colitis. Gut 72, 1451–1461 (2023).
Koch, S. et al. The Wnt antagonist Dkk1 regulates intestinal epithelial homeostasis and wound repair. Gastroenterology 141, 259–268 (2011).
Xu, J. et al. Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO J. 39, e103255 (2020).
Lau, W. de et al. Peyer’s patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured “miniguts. Mol. Cell Biol. 32, 3639–3647 (2012).
Qi, Z. et al. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017).
Li, Q. et al. Interferon-γ and tumor necrosis factor-α disrupt epithelial barrier function by altering lipid composition in membrane microdomains of tight junction. Clin. Immunol. 126, 67–80 (2008).
Reinecker, H. C. et al. Enhanced secretion of tumour necrosis factor-alpha, IL-6, and IL-1 beta by isolated lamina propria mononuclear cells from patients with ulcerative colitis and Crohn’s disease. Clin. Exp. Immunol. 94, 174–181 (1993).
Wirtz, S. & Neurath, M. F. Illuminating the role of type I IFNs in colitis. JCI 115, 586–588 (2005).
Li, J.-Y. et al. IRF/Type I IFN signaling serves as a valuable therapeutic target in the pathogenesis of inflammatory bowel disease. Int. Immunopharmacol. 92, 107350 (2021).
Giuffrida, P., Caprioli, F., Facciotti, F. & Sabatino, A. D. The role of interleukin-13 in chronic inflammatory intestinal disorders. Autoimmun. Rev. 18, 549–555 (2019).
Miyoshi, H., Ajima, R., Luo, C. T., Yamaguchi, T. P. & Stappenbeck, T. S. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113 (2012).
Xie, Z. et al. Recent developments on BMPs and their antagonists in inflammatory bowel diseases. Cell Death Discov. 9, 210 (2023)
Sanchez-Duffhues, G., Williams, E., Goumans, M.-J., Heldin, C.-H. & Dijke, P. T. Bone morphogenetic protein receptors: Structure, function and targeting by selective small molecule kinase inhibitors. Bone 138, 115472 (2020).
Kobayashi, S. et al. Collagen type I-mediated mechanotransduction controls epithelial cell fate conversion during intestinal inflammation. Inflammation and Regeneration 42, (2022).
Fujii, M. et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–793.e6 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Childs, C. J. et al. EPIREGULIN creates a developmental niche for spatially organized human intestinal enteroids. JCI Insight 8, e165566 (2023).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17, 615–620 (2020).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2023).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280.e26 (2021).
Xu, C. et al. Probabilistic harmonization and annotation ofsingle-cell transcriptomics data with deepgenerative models. Mol. Syst. Biol. 17, e9620 (2021).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Ahlmann-Eltze, C. & Huber, W. glmGamPoi: fitting Gamma-Poisson generalized linear models on single cell count data. Bioinformatics 36, 5701–5702 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8, e43803 (2019).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Kuleshov, M. V. et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 44, W90–W97 (2016).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nature. Communications 12, 1088 (2021).
Lin, X., Chau, C., Ma, K., Huang, Y. & Ho, J. W. K. DCATS: differential composition analysis for flexible single-cell experimental designs. Genome Biol. 24, 151 (2023).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2021).
Acknowledgements
We are grateful for the cooperation of Donor Network West and all of the organ and tissue donors and their families, for giving the gift of life and the gift of knowledge by their generous donation. Additionally, we are thankful for Leslie Gaffney at the Broad Research Communication Lab for advice and editing of figures, and for Neko Ota for his help with organoid irradiation. Schematics used in this manuscript were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
M.M.C. and M.B.C. conceived the study. M.M.C. performed all experiments. M.M.C., M.B.C., B.C., K.A., and R.W. analyzed all experiments. E.P., V.I.L., C.F., S.R., and L.M. performed all spatial transcriptomics experiments. M.M.C., L.H., D.P., E.S., M.K., and M.B.C. contributed to organoid generation from human colon tissue and generation of organoid and tissue scRNA-seq data. X.T. and J.L. conceived of organoid cytomix treatment. M.M.C. and C.S. performed organoid microscopy. M.M.C. and M.B.C. wrote the manuscript and produced the figures. All authors edited the manuscript. S.C., L.M., M.K., Z.M., and O.R.R. supervised the work.
Corresponding author
Ethics declarations
Competing interests
All authors are or were employed by Genentech, Inc., South San Francisco, California, at the time of their contribution to this work. M.M.C., B.C., E.P., V.I.L., C.F., S.R., L.H., X.T., D.P., J.L., S.C., E.S., Z.M., L.M., R.W., M.K., O.R.R., and M.B.C. are equity holders in Roche.
Peer review
Peer review information
Nature Communications thanks Toshiro Sato and Shiro Yui for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Capeling, M.M., Chen, B., Aliar, K. et al. Dictionary of human intestinal organoid responses to secreted niche factors at single cell resolution. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68247-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68247-6