Abstract
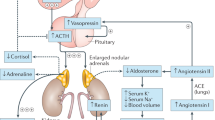
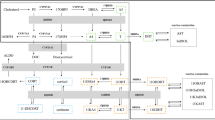
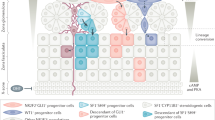
Glucocorticoid-producing cells of the adrenal cortex (i.e. zona fasciculata, zF) constitute the critical effectors of the hypothalamic-pituitary-adrenal axis, mediating the mammalian stress response. With glucocorticoids being essential for life, zF dysfunction perturbs multiple organs that participate in optimizing cardiometabolic fitness. The zF forms a dynamic and heterogenous cell population endowed with the capacity to remodel through the engagement of both proliferative and differentiation programs that enable the adrenal to adapt and respond to diverse stressors. However, the mechanisms that sustain such differential responsiveness remain poorly understood. In this study, we resolve the transcriptome of the steroidogenic lineage by scRNA-seq using Sf1-Crehigh; RosamT/mG reporter mice. We identify HHEX, a homeodomain protein, as the most enriched transcription factor in glucocorticoid-producing cells. We utilize genetic mouse models to demonstrate that Hhex deletion causes glucocorticoid deficiency in male animals. Molecularly, we demonstrate that HHEX is an androgen receptor (AR) target gene, shaping the sexual dimorphism of the adrenal gland by repressing the female transcriptional program at puberty, while also maintaining zF cholesterol ester content by protecting lipid droplets from androgen-induced-lipophagy. Moreover, our study reveals that, in both sexes, HHEX is crucial for maintaining the identity of the innermost adrenocortical cell subpopulation. Specifically, loss of HHEX impairs the expression of Abcb1b (P-glycoprotein/MDR1), an efflux pump regulating steroid export and cellular levels of xenobiotics. Together, these data demonstrate that HHEX serves as a multi-functional regulator of post-natal adrenal maturation that is potentiated by androgens.
Similar content being viewed by others
Data availability
All sequencing datasets generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE291343, GSE291472, and GSE291344. Processed data are provided in Supplementary Data 1. The CLOUPE file related to the scRNAseq dataset presented in this manuscript is accessible at the following address [https://doi.org/10.6084/m9.figshare.28612406]. Source data are provided with this paper.
References
Kuo, T., McQueen, A., Chen, T.-C. & Wang, J.-C. Regulation of glucose homeostasis by glucocorticoids. Adv. Exp. Med. Biol. 872, 99–126 (2015).
Cain, D. W. & Cidlowski, J. A. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 17, 233–247 (2017).
Dickmeis, T. Glucocorticoids and the circadian clock. J. Endocrinol. 200, 3–22 (2009).
Marin, M.-F. et al. Chronic stress, cognitive functioning and mental health. Neurobiol. Learn. Mem. 96, 583–595 (2011).
Shimba, A. & Ikuta, K. Glucocorticoids regulate circadian rhythm of innate and adaptive immunity. Front. Immunol. 11, 2143 (2020).
Lacroix, A. Cardiometabolic morbidity of mild cortisol excess. Ann. Intern. Med. https://doi.org/10.7326/M21-4526 (2022).
Mitani, F. et al. Cytochrome P-45011 beta and P-450scc in adrenal cortex: zonal distribution and intramitochondrial localization by the horseradish peroxidase-labeled antibody method. J. Histochem. Cytochem. 30, 1066–1074 (1982).
Sugano, S. et al. Monoclonal antibodies against bovine adrenal cytochrome P-450(11 beta) and cytochrome P-450SCC. Their isolation, characterization and application to immunohistochemical analysis of adrenal cortex. J. Steroid Biochem. 23, 1013–1021 (1985).
Ogishima, T., Suzuki, H., Hata, J., Mitani, F. & Ishimura, Y. Zone-specific expression of aldosterone synthase cytochrome P-450 and cytochrome P-45011 beta in rat adrenal cortex: histochemical basis for the functional zonation. Endocrinology 130, 2971–2977 (1992).
Ho, M. M. & Vinson, G. P. 11 beta-hydroxylase gene expression in the rat adrenal cortex. J. Endocrinol. 139, 301–306 (1993).
Erdmann, B., Denner, K., Gerst, H., Lenz, D. & Bernhardt, R. Human adrenal CYP11B1: localization by in situ-hybridization and functional expression in cell cultures. Endocr. Res. 21, 425–435 (1995).
Gomez-Sanchez, C. E. et al. Development of monoclonal antibodies against human CYP11B1 and CYP11B2. Mol. Cell. Endocrinol. 383, 111–117 (2014).
Arakane, F. et al. Phosphorylation of steroidogenic acute regulatory protein (StAR) modulates its steroidogenic activity. J. Biol. Chem. 272, 32656–32662 (1997).
Clark, B. J. et al. Hormonal and developmental regulation of the steroidogenic acute regulatory protein. Mol. Endocrinol. Baltim. Md 9, 1346–1355 (1995).
Lopez, J. P. et al. Single-cell molecular profiling of all three components of the HPA axis reveals adrenal ABCB1 as a regulator of stress adaptation. Sci. Adv. 7, eabe4497 (2021).
Lerario, A. M., Mohan, D. R. & Hammer, G. D. Update on biology and genomics of adrenocortical carcinomas: rationale for emerging therapies. Endocr. Rev. 43, 1051–1073 (2022).
Paz, H., Lynch, M. R., Bogue, C. W. & Gasson, J. C. The homeobox gene Hhex regulates the earliest stages of definitive hematopoiesis. Blood 116, 1254–1262 (2010).
Guiral, M., Bess, K., Goodwin, G. & Jayaraman, P. S. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J. Biol. Chem. 276, 2961–2970 (2001).
Swingler, T. E., Bess, K. L., Yao, J., Stifani, S. & Jayaraman, P.-S. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J. Biol. Chem. 279, 34938–34947 (2004).
Yang, D. et al. CRISPR screening uncovers a central requirement for HHEX in pancreatic lineage commitment and plasticity restriction. Nat. Cell Biol. 24, 1064–1076 (2022).
Bort, R. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 131, 797–806 (2004).
Bort, R., Signore, M., Tremblay, K., Barbera, J. P. M. & Zaret, K. S. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 290, 44–56 (2006).
Keng, V. W. et al. Homeobox gene hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem. Biophys. Res. Commun. 276, 1155–1161 (2000).
Martinez Barbera, J. P. et al. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Dev. Camb. Engl. 127, 2433–2445 (2000).
Zhang, J., McKenna, L. B., Bogue, C. W. & Kaestner, K. H. The diabetes gene Hhex maintains -cell differentiation and islet function. Genes Dev. 28, 829–834 (2014).
Alfaifi, M. Contribution of genetic variant identified in HHEX gene in the overweight Saudi patients confirmed with type 2 diabetes mellitus. Saudi J. Biol. Sci. 29, 804–808 (2022).
Wang, X. et al. The association between HHEX single-nucleotide polymorphism rs5015480 and gestational diabetes mellitus: a meta-analysis. Medicine (Baltimore) 99, e19478 (2020).
Li, C. et al. Association between single nucleotide polymorphisms in CDKAL1 and HHEX and type 2 diabetes in Chinese population. Diabetes Metab. Syndr. Obes. Targets Ther. 13, 5113–5123 (2020).
Ragvin, A. et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc. Natl. Acad. Sci. USA 107, 775–780 (2010).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Cauchi, S. et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS ONE 3, e2031 (2008).
van Vliet-Ostaptchouk, J. V. et al. HHEX gene polymorphisms are associated with type 2 diabetes in the Dutch Breda cohort. Eur. J. Hum. Genet. 16, 652–656 (2008).
Chiang, C.-W., Chou, Y.-H., Huang, C.-N., Lu, W.-Y. & Liaw, Y.-P. Gender-specific genetic influence of rs1111875 on diabetes risk: insights from the Taiwan biobank study. J. Diabetes Investig. https://doi.org/10.1111/jdi.14359 (2024).
Zhai, G. et al. Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet. 7, e1002025 (2011).
Vernerova, L. et al. Contribution of genetic factors to lower DHEAS in patients with rheumatoid arthritis. Cell. Mol. Neurobiol. 38, 379–383 (2018).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Bingham, N. C., Verma-Kurvari, S., Parada, L. F. & Parker, K. L. Development of a steroidogenic factor 1/Cre transgenic mouse line. Genes 44, 419–424 (2006).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Basham, K. J. et al. A ZNRF3-dependent Wnt/β-catenin signaling gradient is required for adrenal homeostasis. Genes Dev. 33, 209–220 (2019).
Mathieu, M. et al. Steroidogenic differentiation and PKA signaling are programmed by histone methyltransferase EZH2 in the adrenal cortex. Proc. Natl. Acad. Sci. USA 115, E12265–E12274 (2018).
Dufour, D. et al. Loss of SUMO-specific protease 2 causes isolated glucocorticoid deficiency by blocking adrenal cortex zonal transdifferentiation in mice. Nat. Commun. 13, 7858 (2022).
Gjerstad, J. K., Lightman, S. L. & Spiga, F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 21, 403–416 (2018).
Ramamoorthy, S. & Cidlowski, J. A. Corticosteroids. Rheum. Dis. Clin. N. Am. 42, 15–31 (2016).
Holst, J. P., Soldin, O. P., Guo, T. & Soldin, S. J. Steroid hormones: relevance and measurement in the clinical laboratory. Clin. Lab. Med. 24, 105–118 (2004).
Rosol, T. J. & Gröne, A. Chapter 3 - Endocrine glands. In Jubb, Kennedy & Palmer’s Pathology of Domestic Animals: Volume 3 (Sixth Edition) (ed. Maxie, M. G.) 269–357.e1 (W.B. Saunders, 2016).
Acconcia, F. & Marino, M. Steroid hormones: synthesis, secretion, and transport. In Principles of Endocrinology and Hormone Action (eds Belfiore, A. & LeRoith, D.) 43–72 (Springer International Publishing, Cham, 2018).
Lightman, S. L., Birnie, M. T. & Conway-Campbell, B. L. Dynamics of ACTH and cortisol secretion and implications for disease. Endocr. Rev. 41, bnaa002 (2020).
Dailey, R. E., Swell, L. & Treadwell, C. R. Utilization of free and esterified gholesterol-4-G14 for corticoid biosynthesis by hog adrenal homogenates. Proc. Soc. Exp. Biol. Med. 110, 571–574 (1962).
Long, C. N. H. The relation of cholesterol and ascorbic acid to the secretion of the adrenal cortex. Recent Prog. Horm. Res. 1, 99–122 (1947).
Wang, N., Wang, W., Breslow, J. L. & Tall, A. R. Scavenger receptor BI (SR-BI) is up-regulated in adrenal gland in apolipoprotein A-I and hepatic lipase knock-out mice as a response to depletion of cholesterol stores. In vivo evidence that SR-BI is a functional high density lipoprotein receptor under feedback control. J. Biol. Chem. 271, 21001–21004 (1996).
Shroff, A. & Nazarko, T. Y. SQSTM1, lipid droplets and current state of their lipophagy affairs. Autophagy 19, 720–723 (2023).
Wang, L. et al. Ethanol-triggered lipophagy requires SQSTM1 in AML12 hepatic cells. Sci. Rep. 7, 12307 (2017).
Kumar, A. V., Mills, J. & Lapierre, L. R. Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front. Cell Dev. Biol. 10, 793328 (2022).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Freedman, B. D. et al. Adrenocortical zonation results from lineage conversion of differentiated zona glomerulosa cells. Dev. Cell 26, 666–673 (2013).
Pihlajoki, M. et al. Conditional mutagenesis of Gata6 in SF1-positive cells causes gonadal-like differentiation in the adrenal cortex of mice. Endocrinology https://doi.org/10.1210/en.2012-1892 (2013).
Mauthe, M. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018).
Beyer, C. & Komisaruk, B. Effects of diverse androgens on estrous behavior, lordosis reflex, and genital tract morphology in the rat. Horm. Behav. 2, 217–225 (1971).
Brown-Grant, K., Munck, A., Naftolin, F. & Sherwood, M. R. The effects of the administration of testosterone propionate alone or with phenobarbitone and of testosterone metabolites to neonatal female rats. Horm. Behav. 2, 173–182 (1971).
De Gendt, K. et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl. Acad. Sci. USA. 101, 1327–1332 (2004).
Nikkanen, J. et al. An evolutionary trade-off between host immunity and metabolism drives fatty liver in male mice. Science 378, 290–295 (2022).
Jo, S. et al. Sex differences in pancreatic β-cell physiology and glucose homeostasis in C57BL/6J mice. J. Endocr. Soc. 7, bvad099 (2023).
Dumontet, T. et al. PKA signaling drives reticularis differentiation and sexually dimorphic adrenal cortex renewal. JCI Insight 3, e98394 (2018).
Grabek, A. et al. The adult adrenal cortex undergoes rapid tissue renewal in a sex-specific manner. Cell Stem Cell 25, 290–296.e2 (2019).
Levasseur, A., Dumontet, T. & Martinez, A. “Sexual dimorphism in adrenal gland development and tumorigenesis”. Curr. Opin. Endocr. Metab. Res. 8, 60–65 (2019).
Yeung, K. Y. & Ruzzo, W. L. Principal component analysis for clustering gene expression data. Bioinforma. Oxf. Engl. 17, 763–774 (2001).
Lyu, Q. et al. RNA-seq reveals sub-zones in mouse adrenal zona fasciculata and the sexually dimorphic responses to thyroid hormone. Endocrinology 161, bqaa126 (2020).
Wakil, A. E., Mari, B., Barhanin, J. & Lalli, E. Genomic analysis of sexual dimorphism of gene expression in the mouse adrenal gland. Horm. Metab. Res. 45, 870–873 (2013).
Mukai, T. et al. Sexually dimorphic expression of Dax-1 in the adrenal cortex. Genes Cells Devoted Mol. Cell. Mech. 7, 717–729 (2002).
Devine, K. et al. The ATP-binding cassette proteins ABCB1 and ABCC1 as modulators of glucocorticoid action. Nat. Rev. Endocrinol. 19, 112–124 (2023).
Menzies, R. I. et al. Transcription controls growth, cell kinetics and cholesterol supply to sustain ACTH responses. Endocr. Connect. 6, 446–457 (2017).
Vogel, F. et al. Polymorphism in the drug transporter gene ABCB1 as a potential disease modifier in cortisol-producing adrenal adenomas. Exp. Clin. Endocrinol. Diabetes 132, 608–613 (2024).
Hammer, G. D. & Basham, K. J. Stem cell function and plasticity in the normal physiology of the adrenal cortex. Mol. Cell. Endocrinol. 519, 111043 (2021).
Guasti, L., Paul, A., Laufer, E. & King, P. Localization of Sonic hedgehog secreting and receiving cells in the developing and adult rat adrenal cortex. Mol. Cell. Endocrinol. 336, 117–122 (2011).
Neirijnck, Y. et al. Single-cell transcriptomic profiling redefines the origin and specification of early adrenogonadal progenitors. Cell Rep. 42, 112191 (2023).
Pivovarova, O., Nikiforova, V. J., Pfeiffer, A. F. H. & Rudovich, N. The influence of genetic variations in HHEX gene on insulin metabolism in the German MESYBEPO cohort. Diabetes Metab. Res. Rev. 25, 156–162 (2009).
Staiger, H. et al. A candidate type 2 diabetes polymorphism near the HHEX locus affects acute glucose-stimulated insulin release in European populations: results from the EUGENE2 study. Diabetes 57, 514–517 (2008).
Pascoe, L. et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes 56, 3101–3104 (2007).
Grarup, N. et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 danish subjects: validation and extension of genome-wide association studies. Diabetes 56, 3105–3111 (2007).
Liu, S. et al. Genetic variants at 10q23.33 are associated with plasma lipid levels in a Chinese population. J. Biomed. Res. 28, 53–58 (2014).
Dumontet, T. & Martinez, A. Adrenal androgens, adrenarche, and zona reticularis: a human affair? Mol. Cell. Endocrinol. 528, 111239 (2021).
Kraemer, F. B. et al. Adrenal neutral cholesteryl ester hydrolase: identification, subcellular distribution, and sex differences. Endocrinology 143, 801–806 (2002).
Li, H. et al. Hormone-sensitive lipase deficiency in mice causes lipid storage in the adrenal cortex and impaired corticosterone response to corticotropin stimulation. Endocrinology 143, 3333–3340 (2002).
Al, E. M. et al. Wolman’s disease: the king faisal specialist hospital and research centre experience. Ann. Saudi Med. 18, 120–124 (1998).
Perry, R. et al. Primary adrenal insufficiency in children: twenty years experience at the Sainte-Justine Hospital, Montreal. J. Clin. Endocrinol. Metab. 90, 3243–3250 (2005).
Menon, J. et al. Wolman’s disease: a rare cause of infantile cholestasis and cirrhosis. J. Pediatr. Genet. 11, 132–134 (2020).
Foladi, N. & Aien, M. T. CT features of Wolman disease (lysosomal acid lipase enzyme deficiency) – A case report. Radiol. Case Rep. 16, 2857–2861 (2021).
Sen, D., Satija, L., Saxena, S., Rastogi, V. & Singh, M. A rare constellation of imaging findings in Wolman disease. Med. J. Armed Forces India 71, S448–S451 (2015).
Fulcher, A. S., Das Narla, L. & Hingsbergen, E. A. Pediatric case of the day. Wolman disease (primary familial xanthomatosis with involvement and calcification of the adrenal glands). RadioGraphics 18, 533–535 (1998).
Wolman, M., Sterk, V. V., Gatt, S. & Frenkel, M. Primary familial xanthomatosis with involvement and calcification of the adrenals. Report of two more cases in siblings of a previously described infant. Pediatrics 28, 742–757 (1961).
Abramov, A., Schorr, S. & Wolman, M. Generalized xanthomatosis with calcified adrenals. AMA J. Dis. Child. 91, 282–286 (1956).
Low, G., Irwin, G. J., MacPhee, G. B. & Robinson, P. H. Characteristic imaging findings in Wolman’s disease. Clin. Radiol. Extra 59, 106–108 (2004).
Schaub, J. et al. Wolman’s disease: clinical, biochemical and ultrastructural studies in an unusual case without striking adrenal calcification. Eur. J. Pediatr. 135, 45–53 (1980).
Zhang, S. et al. The regulation, function, and role of lipophagy, a form of selective autophagy, in metabolic disorders. Cell Death Dis. 13, 1–11 (2022).
Gao, F. et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J. Cell Biol. 217, 2103–2119 (2018).
Ma, Y. et al. Lipophagy contributes to testosterone biosynthesis in male rat leydig cells. Endocrinology 159, 1119–1129 (2018).
Esmaeilian, Y. et al. Autophagy regulates sex steroid hormone synthesis through lysosomal degradation of lipid droplets in human ovary and testis. Cell Death Dis. 14, 1–13 (2023).
Berruti, A. et al. Prognostic role of overt hypercortisolism in completely operated patients with adrenocortical cancer. Eur. Urol. 65, 832–838 (2014).
Vanbrabant, T., Fassnacht, M., Assie, G. & Dekkers, O. M. Influence of hormonal functional status on survival in adrenocortical carcinoma: systematic review and meta-analysis. Eur. J. Endocrinol. 179, 429–436 (2018).
Jeong, S.-J. et al. Prdx1 (peroxiredoxin 1) deficiency reduces cholesterol efflux via impaired macrophage lipophagic flux. Autophagy 14, 120–133 (2018).
Chen, K., Yuan, R., Zhang, Y., Geng, S. & Li, L. Tollip deficiency alters atherosclerosis and steatosis by disrupting lipophagy. J. Am. Heart Assoc. 6, e004078 (2017).
Ouimet, M. et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 37, 1058–1067 (2017).
Carotti, S. et al. Lipophagy impairment is associated with disease progression in NAFLD. Front. Physiol. 11, 850 (2020).
Fu, Y. et al. Degradation of lipid droplets by chimeric autophagy-tethering compounds. Cell Res. 31, 965–979 (2021).
Minami, Y. et al. Liver lipophagy ameliorates nonalcoholic steatohepatitis through extracellular lipid secretion. Nat. Commun. 14, 4084 (2023).
Müller, M. B. et al. ABCB1 (MDR1)-type P-glycoproteins at the blood-brain barrier modulate the activity of the hypothalamic-pituitary-adrenocortical system: implications for affective disorder. Neuropsychopharmacology 28, 1991–1999 (2003).
Flynn, S. D. et al. P-glycoprotein expression and multidrug resistance in adrenocortical carcinoma. Surgery 112, 981–986 (1992).
Creemers, S. G. et al. MDR1 inhibition increases sensitivity to doxorubicin and etoposide in adrenocortical cancer. Endocr. Relat. Cancer 26, 367–378 (2019).
Bechmann, N. et al. Asymmetric adrenals: sexual dimorphism of adrenal tumors. J. Clin. Endocrinol. Metab. 109, 471–482 (2024).
Takahashi, F. et al. Development of sexual dimorphism of skeletal muscles through the adrenal cortex, caused by androgen-induced global gene suppression. Cell Rep. 43, 113715 (2024).
Wilson, S., Qi, J. & Filipp, F. V. Refinement of the androgen response element based on ChIP-Seq in androgen-insensitive and androgen-responsive prostate cancer cell lines. Sci. Rep. 6, 32611 (2016).
Hunter, M. P. et al. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev. Biol. 308, 355–367 (2007).
Ferreira, M. J. et al. Spontaneous pancreatitis caused by tissue-specific gene ablation of Hhex in mice. Cell. Mol. Gastroenterol. Hepatol. 1, 550–569 (2015).
Watanabe, H. et al. Transcription factor hematopoietically expressed homeobox protein (Hhex) negatively regulates osteoclast differentiation by controlling cyclin-dependent kinase inhibitors. JBMR Plus 6, e10608 (2022).
Jackson, J. T. et al. A crucial role for the homeodomain transcription factor Hhex in lymphopoiesis. Blood 125, 803–814 (2015).
Kim, A. C. et al. Targeted disruption of β-catenin in Sf1-expressing cells impairs development and maintenance of the adrenal cortex. Development 135, 2593–2602 (2008).
King, P., Paul, A. & Laufer, E. Shh signaling regulates adrenocortical development and identifies progenitors of steroidogenic lineages. Proc. Natl. Acad. Sci. USA 106, 21185–21190 (2009).
Huang, C.-C. J. & Yao, H. H. Inactivation of Dicer1 in Steroidogenic factor 1-positive cells reveals tissue-specific requirement for Dicer1 in adrenal, testis, and ovary. BMC Dev. Biol. 10, 66 (2010).
Tevosian, S. G. et al. Adrenal development in mice requires GATA4 and GATA6 transcription factors. Endocrinology 156, 2503–2517 (2015).
Drelon, C. et al. PKA inhibits WNT signalling in adrenal cortex zonation and prevents malignant tumour development. Nat. Commun. 7, 12751 (2016).
Vidal, V. et al. The adrenal capsule is a signaling center controlling cell renewal and zonation through Rspo3. Genes Dev. 30, 1389–1394 (2016).
Dumontet, T. et al. Hormonal and spatial control of SUMOylation in the human and mouse adrenal cortex. FASEB J. 33, 10218–10230 (2019).
Heaton, J. H. et al. Progression to adrenocortical tumorigenesis in mice and humans through insulin-like growth factor 2 and β-catenin. Am. J. Pathol. 181, 1017–1033 (2012).
Krill, K. T., Gurdziel, K., Heaton, J. H., Simon, D. P. & Hammer, G. D. Dicer deficiency reveals microRNAs predicted to control gene expression in the developing adrenal cortex. Mol. Endocrinol. 27, 754–768 (2013).
Ching, S. & Vilain, E. Targeted disruption of Sonic Hedgehog in the mouse adrenal leads to adrenocortical hypoplasia. Genesis 47, 628–637 (2009).
Truett, G. E. et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29, 52 (2000).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Hippen, A. A. et al. miQC: an adaptive probabilistic framework for quality control of single-cell RNA-sequencing data. PLoS Comput. Biol. 17, e1009290 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Alquicira-Hernandez, J. & Powell, J. E. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinforma. Oxf. Engl. 37, 2485–2487 (2021).
Cummins, C. L. et al. Liver X receptors regulate adrenal cholesterol balance. J. Clin. Invest. 116, 1902–1912 (2006).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Rueden, C. T. et al. ImageJ2: imageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinforma. Oxf. Engl. 28, 882–883 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Acknowledgements
Research reported in this publication was supported by the International Fund for Congenital Adrenal Hyperplasia to G.D.H., F.B., and T.D., the Center for Cell Plasticity and Organ Design to T.D., the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number R01DK043140, R01DK062027 to G.D.H. and Dr. William Rainey, the National Heart, Lung, and Blood Institute under Award Number 1R01HL15583401 to A.F.T., the Swiss National Science Foundation (310030L_182700/1) to F.B., the Swiss NCCR “Kidney.CH” and from the University Research Priority Program of the University of Zurich ITINERARE–Innovative Therapies in Rare Disease to D.P. and F.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank all the members of the Hammer Lab for helpful discussion and feedback on the manuscript. We especially thank Dr. Rainey (University of Michigan) for providing Human adrenal paraffin samples and sharing ARKO RNAseq. We thank Dr. Pierre Val (University of Clermont-Ferrand) for sharing reagents. We thank Dr. Gregg Myers (University of Michigan) for technical assistance with hematopoietic cells. We thank Dr. Leonard Cheung (Stony Brook University) for helpful discussion regarding HHEX expression in the mouse pituitary gland. We thank Dr. Klionsky (University of Michigan), autophagy expert for helpful discussion on lipophagy process. We thank Dr. Frank Claessens and Dr. Johan Swinnen (KU Leuven) for sharing the AR flox mice60. The research reported in this publication used the Advanced Genomic Core, the Microscopy Core, the Flow Cytometry Core, and the Orthopaedic Research Laboratories (ORL) Histology Core at the University of Michigan. We especially thank Emma Snyder-White and Carol Whitinger for technical assistance with histology experiments and Michael Pihalja with FACS sorting.
Author information
Authors and Affiliations
Contributions
We followed guidelines for authorship at University of Michigan Office of Research including 4 criteria. (1) significant contribution to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; (2) drafting the work or revising it critically for important intellectual content; (3) final approval of the version to be published; (4) agreement to be accountable for all aspects of the work in ensuring that questions related to accuracy or integrity of any part of the work are appropriately investigated. T.D., K.J.B., and G.D.H. design the experiments, T.D. and A.M.L. analyzed the data, T.D., K.J.B., M.C.F., C.R.L., D.J, E.S., and K.A.H. genotyped mice, performed the experiments, or acquired data. C.L. and A.F.T. performed LCMSMS. S.W.P. and A.M.L. analyzed the ARKO RNAseq data. D.P. and F.B. provided Cyp11b1-CreERT2 mice. D.T.B. provided Cyp11b2-Cre (AS-Cre) mice. T.D. wrote the original manuscript, T.D., K.J.B., C.R.L., and G.D.H. edited the manuscript. All coauthors provided expertise and feedback.
Corresponding author
Ethics declarations
Competing interests
G.D.H. Founder and Board of Directors—Sling Therapeutics, Advisor—Orphagen Pharmaceuticals. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dumontet, T., Basham, K.J., Foster, M.C. et al. The transcription factor HHEX maintains glucocorticoid levels and protects adrenals from androgen-induced lipid depletion. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68257-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68257-4