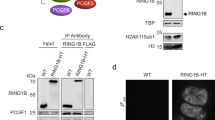
Abstract
Polycomb repressive complex 2 (PRC2) mediates developmental gene repression as two classes of holocomplexes, PRC2.1 and PRC2.2. EPOP is an accessory subunit specific to PRC2.1, which also contains PCL proteins. Unlike other accessory subunits that collectively facilitate PRC2 targeting, EPOP was implicated in an enigmatic inhibitory role, together with its interactor Elongin BC. We report an unusual molecular mechanism whereby EPOP regulates PRC2.1 by directly modulating its oligomerization state. EPOP disrupts the PRC2.1 dimer and weakens its chromatin association, likely by disabling the avidity effect conferred by the dimeric complex. Congruently, an EPOP mutant specifically defective in PRC2 binding enhances genome-wide enrichments of MTF2 in mouse epiblast-like cells. Elongin BC is largely dispensable for the EPOP-mediated inhibition of PRC2.1. EPOP defines a distinct subclass of PRC2.1, which may uniquely maintain an epigenetic program by preventing the over-repression of key gene regulators along the continuum of early differentiation.
Similar content being viewed by others
Data availability
The crystal structure described in this work has been deposited in the Protein Data Bank (PDB) under the accession number 9XZI. Genomics data have been deposited in NCBI’s Gene Expression Omnibus under the accession numbers GSE264672 and GSE264673. LC-MS/MS data files have been deposited to the ProteomeXchange via the MassIVE partner repository with the accession code PXD072233. Source data are provided in this paper.
References
Schuettengruber, B., Bourbon, H. M., Di Croce, L. & Cavalli, G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 (2017).
Comet, I., Riising, E. M., Leblanc, B. & Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803–810 (2016).
Grijzenhout, A. et al. Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 143, 2716–2723 (2016).
Liu, X. & Liu, X. PRC2, Chromatin regulation, and human disease: insights from molecular structure and function. Front. Oncol. 12, 894585 (2022).
Hojfeldt, J. W. et al. Non-core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol. Cell 76, 423–436 (2019).
Healy, E. et al. PRC2.1 and PRC2.2 Synergize to coordinate H3K27 trimethylation. Mol. Cell 76, 437–452 (2019).
Petracovici, A. & Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 81, 2625–2639 (2021).
Asenjo, H. G. et al. Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Sci. Adv. 6, eaay4768 (2020).
Vaziri, A. et al. Persistent epigenetic reprogramming of sweet taste by diet. Sci. Adv. 6, https://doi.org/10.1126/sciadv.abc8492 (2020).
Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Jiao, L. & Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015).
Gong, L. et al. CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site. Nat. Commun. 13, 6781 (2022).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Justin, N. et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 (2016).
Ragazzini, R. et al. EZHIP constrains polycomb repressive complex 2 activity in germ cells. Nat. Commun. 10, 3858 (2019).
Jain, S. U. et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 10, 2146 (2019).
Hubner, J. M. et al. EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol. 21, 878–889 (2019).
Piunti, A. et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 5, eaax2887 (2019).
Harutyunyan, A. S. et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun. 10, 1262 (2019).
Jain, S. U. et al. H3 K27M and EZHIP Impede H3K27-Methylation Spreading by Inhibiting Allosterically Stimulated PRC2. Mol. Cell 80, 726–735 (2020).
Kalb, R. et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 (2014).
Cooper, S. et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 (2016).
Kasinath, V. et al. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371, https://doi.org/10.1126/science.abc3393 (2021).
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 (2014).
Cooper, S. et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470 (2014).
Li, H. et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291 (2017).
Perino, M. et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 50, 1002–1010 (2018).
Ballare, C. et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 19, 1257–1265 (2012).
Musselman, C. A. et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 19, 1266–1272 (2012).
Brien, G. L. et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 19, 1273–1281 (2012).
Cai, L. et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 49, 571–582 (2013).
Chen, S., Jiao, L., Liu, X., Yang, X. & Liu, X. A dimeric structural scaffold for PRC2-PCL targeting to CpG Island chromatin. Mol. Cell 77, 1265–1278 (2020).
Amoutzias, G. D., Robertson, D. L., Van de Peer, Y. & Oliver, S. G. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 33, 220–229 (2008).
Zhang, Z. et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29, 229–240 (2011).
De Cegli, R. et al. Reverse engineering a mouse embryonic stem cell-specific transcriptional network reveals a new modulator of neuronal differentiation. Nucleic Acids Res. 41, 711–726 (2013).
Smits, A. H., Jansen, P. W., Poser, I., Hyman, A. A. & Vermeulen, M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 41, e28 (2013).
Alekseyenko, A. A., Gorchakov, A. A., Kharchenko, P. V. & Kuroda, M. I. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. USA 111, 2488–2493 (2014).
Liefke, R. & Shi, Y. The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discov. 1, 15008 (2015).
Liefke, R., Karwacki-Neisius, V. & Shi, Y. EPOP Interacts with Elongin BC and USP7 to modulate the chromatin landscape. Mol. Cell 64, 659–672 (2016).
Beringer, M. et al. EPOP Functionally links elongin and polycomb in pluripotent stem cells. Mol. Cell 64, 645–658 (2016).
Kloet, S. L. et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 23, 682–690 (2016).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Chen, S., Jiao, L., Shubbar, M., Yang, X. & Liu, X. Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol. Cell 69, 840–852 e5 (2018).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Meszaros, B., Erdos, G. & Dosztanyi, Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018).
Hauri, S. et al. A high-density map for navigating the human polycomb complexome. Cell Rep. 17, 583–595 (2016).
Rougeot, J., Renard, M., Randsholt, N. B., Peronnet, F. & Mouchel-Vielh, E. The elongin complex antagonizes the chromatin factor Corto for vein versus intervein cell identity in Drosophila wings. PLoS ONE 8, e77592 (2013).
Kamura, T. et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12, 3872–3881 (1998).
Schoenfeld, A. R., Davidowitz, E. J. & Burk, R. D. Elongin BC complex prevents degradation of von Hippel-Lindau tumor suppressor gene products. Proc. Natl. Acad. Sci. USA 97, 8507–8512 (2000).
Dennis, G. Jr. et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 (2003).
Gillotin, S. Isolation of chromatin-bound proteins from subcellular fractions for biochemical analysis. Bio. Protoc. 8, e3035 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Bricogne, G. et al. BUSTER version 2.10.4. Cambridge, United Kingdom: Global Phasing Ltd. (2017).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Schrodinger, L. The PyMOL Molecular Graphics System, Version 2.5.5 (2010).
Acknowledgements
The cDNA of EPOP was kindly provided by Dr. Robert Liefke and Dr. Yang Shi. The cDNAs of human PRC2 core subunits were kindly provided by Dr. Robert Kingston. This research was supported by NIH grants R35GM136308 and R21NS137323 to X.L. X.L. is a W.W. Caruth, Jr., Scholar in Biomedical Research. L.G. was supported by an American Heart Association postdoctoral fellowship 19POST34450043, the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB1230000), NSFC-Joint Foundation of Yunnan Province (U24A20806), Introducing Talents Start-up Fund of Kunming Institute of Botany, Chinese Academy of Sciences (51E46G0111K1), and Fund of State Key Laboratory of Phytochemistry and Natural Medicines (52E46P5211Z1). S.C. was supported by the National Natural Science Foundation of China grant 32100464. Some data presented in this report were acquired with a mass photometer that was supported by an award S10OD030312-01 from the NIH. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Author information
Authors and Affiliations
Contributions
Xin.L. conceived the study; L.G., Xiuli.L., M.Y.B, X.Y., S.C., and Xin.L. designed and performed the experiments; X.Y. and Xin.L. analyzed the structural data; Xiuli.L., Z.Y., C.X., and Xin.L. analyzed the genomics data; A.L. analyzed the proteomics data; L.G., Xiuli.L., X.Y., Z.Y., C.X., and Xin.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tatiana Kutateladze and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gong, L., Liu, X., Yang, X. et al. EPOP restricts PRC2.1 targeting to chromatin by directly modulating enzyme complex dimerization. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68280-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68280-5