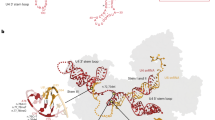
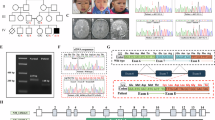
Abstract
SF3B1 is an essential and ubiquitous splicing factor that plays a pivotal role in the early steps of pre-mRNA splicing. Recurrent somatic missense mutations in SF3B1 are frequent in cancers, but no constitutional variant has been reported so far. We describe here a cohort of 26 individuals with neurodevelopmental disorders, harbouring SF3B1 constitutional heterozygous variants that appeared mostly de novo. Patients present with a global developmental delay, associated with variable neurological and facial dysmorphic traits. A dichotomy may emerge between patients harbouring predicted loss of function (n = 9) and missense variants (n = 17), the latter being associated with a more severe and syndromic phenotype, including heart and gastrointestinal anomalies. We focused on de novo SF3B1 missense variants, which were largely distinct from those reported in cancer. Functional complementation assays show that de novo SF3B1 missense variants did not cause a loss of function of the protein. Targeted and genome-wide analysis of RNA splicing reveal that they affect canonical and alternative splicing more moderately than somatic variants, and subtly modify the splicing of many transcripts. These findings place SF3B1 among the rare U2 snRNP components implicated in both cancer and neurodevelopmental disorders, highlighting its critical and multifaceted role in human disease.
Similar content being viewed by others
Data availability
The RNA sequencing data generated from patient-derived lymphocytes have been deposited in the European Genome–Phenome Archive (EGA, http://www.ebi.ac.uk/ega), under accession code EGAS50000001473, and are subjected to a data processing agreement due to their sensitive nature. Access will be provided only for health or medical or biomedical research, only for non commercial use, and a collaboration with the primary study investigator is required. Due to the sensitive nature of human genetic data, controlled access to human genome data is necessary to protect participant privacy and to ensure compliance with ethical and legal standards. Individual genome data provided within a clinical diagnostic setting, which are stored on secure hospital servers, cannot be made available due to ethical and regulatory considerations, including patient consent limitations and data protection regulations. However, whole exome sequencing data performed in the framework of the HUGODIMS consortium, as well as the exome sequencing data generated in a research setting, are available from the corresponding author under controlled access. Access will be restricted to qualified researchers for non-commercial research purposes and subject to a data processing agreement. Data access requests for RNA sequencing and exome sequencing data generated in a research setting should be submitted to the corresponding author and will be reviewed within 30 days by a data accessibility committee to ensure that data access complies with ethical and legal standards respective to the corresponding projects. The RNA sequencing data generated in this study on K562 cells have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds) under accession code GSE287369. All other data supporting the findings of this study are available within the paper and its supplementary information files. The source data underlying Figs. 3 and 5 and Supplementary Fig . 4-6 are provided as a Source Data file. Source data are provided with this paper.
References
Marasco, L. E. & Kornblihtt, A. R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 24, 242–254 (2023).
Bonnal, S. C., López-Oreja, I. & Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—implications for care. Nat. Rev. Clin. Oncol. 17, 457–474 (2020).
Griffin, C. & Saint-Jeannet, J.-P. Spliceosomopathies: Diseases and mechanisms. Dev. Dyn. Publ. Am. Assoc. Anat. 249, 1038–1046 (2020).
Cretu, C. et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Alsafadi, S. et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016).
Darman, R. B. et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3’ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015).
Shiozawa, Y. et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat. Commun. 9, 3649 (2018).
Dolatshad, H. et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 30, 2322–2331 (2016).
Dalton, W. B. et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 4, 1192–1196 (2020).
Choi, I. Y. et al. The E592K variant of SF3B1 creates unique RNA missplicing and associates with high-risk MDS without ring sideroblasts. Blood Adv. 8, 3961–3971 (2024).
El Chehadeh, S. et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur. J. Hum. Genet. EJHG 25, 43–51 (2016).
Wang, Q., Moore, M. J., Adelmant, G., Marto, J. A. & Silver, P. A. PQBP1, a factor linked to intellectual disability, affects alternative splicing associated with neurite outgrowth. Genes Dev. 27, 615–626 (2013).
Yang, Y. et al. Prevalence of neurodevelopmental disorders among US children and adolescents in 2019 and 2020. Front. Psychol. 13, 997648 (2022).
Gidziela, A. et al. A meta-analysis of genetic effects associated with neurodevelopmental disorders and co-occurring conditions. Nat. Hum. Behav. 7, 642–656 (2023).
Gillentine, M. A., Wang, T. & Eichler, E. E. Estimating the prevalence of de novo monogenic neurodevelopmental disorders from large cohort studies. Biomedicines 10, 2865 (2022).
Lagrán, M. M. de, Bascón-Cardozo, K. & Dierssen, M. Neurodevelopmental disorders: 2024 update. Free Neuropathol. 5, 20 (2024).
Manickam, K. et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. J. Am. Coll. Med. Genet. 23, 2029–2037 (2021).
Li, D. et al. Spliceosome malfunction causes neurodevelopmental disorders with overlapping features. J. Clin. Invest. 134, e171235 (2024).
Deutsch, H. M., Song, Y. & Li, D. Spliceosome complex and neurodevelopmental disorders. Curr. Opin. Genet. Dev. 93, 102358 (2025).
Greene, D. et al. Mutations in the small nuclear RNA gene RNU2-2 cause a severe neurodevelopmental disorder with prominent epilepsy. Nat. Genet. 57, 1367–1373 (2025).
Low, K. J. et al. PUF60 variants cause a syndrome of ID, short stature, microcephaly, coloboma, craniofacial, cardiac, renal and spinal features. Eur. J. Hum. Genet. 25, 552–559 (2017).
Dauber, A. et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 93, 798–811 (2013).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
Yeo, G. & Burge, C. B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. J. Comput. Mol. Cell Biol. 11, 377–394 (2004).
Leman, R. et al. SPiP: Splicing prediction pipeline, a machine learning tool for massive detection of exonic and intronic variant effects on mRNA splicing. Hum. Mutat. 43, 2308–2323 (2022).
Jian, X., Boerwinkle, E. & Liu, X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 42, 13534–13544 (2014).
Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e24 (2019).
Yang, F. et al. Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly. Nat. Commun. 14, 897 (2023).
Zhang, X. et al. Structural insights into branch site proofreading by human spliceosome. Nat. Struct. Mol. Biol. 31, 835–845 (2024).
Bergot, T. et al. Human cancer-associated mutations of SF3B1 lead to a splicing modification of its own RNA. Cancers 12, 652 (2020).
Dolatshad, H. et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 29, 1798 (2015).
Corrionero, A., Miñana, B. & Valcárcel, J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 25, 445–459 (2011).
Seiler, M. et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.e4 (2018).
Kesarwani, A. K. et al. Cancer-associated SF3B1 mutants recognize otherwise inaccessible cryptic 3’ splice sites within RNA secondary structures. Oncogene 36, 1123–1133 (2017).
Liberante, F. G. et al. Altered splicing and cytoplasmic levels of tRNA synthetases in SF3B1-mutant myelodysplastic syndromes as a therapeutic vulnerability. Sci. Rep. 9, 2678 (2019).
Florea, L., Song, L. & Salzberg, S. L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2, 188 (2013).
Ikeda, F. et al. Exome sequencing identified RPS15A as a novel causative gene for Diamond-Blackfan anemia. Haematologica 102, e93–e96 (2017).
Tajima, H. et al. Evidence for in vivo production of Humanin peptide, a neuroprotective factor against Alzheimer’s disease-related insults. Neurosci. Lett. 324, 227–231 (2002).
Liu, Z. et al. Mutations in the RNA splicing Factor SF3B1 promote tumorigenesis through MYC stabilization. Cancer Discov. 10, 806–821 (2020).
Hwang, J. Y. et al. rMAPS2: An update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 48, W300–W306 (2020).
Duijkers, F. A. et al. HNRNPR variants that impair homeobox gene expression drive developmental disorders in humans. Am. J. Hum. Genet. 104, 1040–1059 (2019).
de Masfrand, S. et al. Penetrance, variable expressivity and monogenic neurodevelopmental disorders. Eur. J. Med. Genet. 69, 104932 (2024).
Chen, P. et al. Phenotypic spectrum and molecular basis in a chinese cohort of osteogenesis imperfecta with mutations in type I collagen. Front. Genet. 13, 816078 (2022).
Guo, L. et al. Null and missense mutations of ERI1 cause a recessive phenotypic dichotomy in humans. Am. J. Hum. Genet. 110, 1068–1085 (2023).
Chettle, J. et al. LARP1 haploinsufficiency is associated with an autosomal dominant neurodevelopmental disorder. HGG Adv. 100345 https://doi.org/10.1016/j.xhgg.2024.100345 (2024).
Nussinov, R., Tsai, C.-J. & Jang, H. How can same-gene mutations promote both cancer and developmental disorders? Sci. Adv. 8, eabm2059 (2022).
Timberlake, A. T. et al. Haploinsufficiency of SF3B2 causes craniofacial microsomia. Nat. Commun. 12, 4680 (2021).
Bernier, F. P. et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am. J. Hum. Genet. 90, 925–933 (2012).
Sobreira, N., Schiettecatte, F., Valle, D. & Hamosh, A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930 (2015).
SIFT - Predict effects of nonsynonmous / missense variants. https://sift.bii.a-star.edu.sg/.
PolyPhen-2: Prediction of functional effects of human nsSNPs. http://genetics.bwh.harvard.edu/pph2/.
Schwarz, J. M., Rödelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Kim, S., Jhong, J.-H., Lee, J. & Koo, J.-Y. Meta-analytic support vector machine for integrating multiple omics data. BioData Min. 10, 2 (2017).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47, D886–D894 (2019).
Ioannidis, N. M. et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Silk, M., Petrovski, S. & Ascher, D. B. MTR-Viewer: Identifying regions within genes under purifying selection. Nucleic Acids Res. 47, W121–W126 (2019).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. J. Am. Coll. Med. Genet. 17, 405–424 (2015).
Abou Tayoun, A. N. et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 39, 1517–1524 (2018).
Stenton, S. L. et al. Assessment of the evidence yield for the calibrated PP3/BP4 computational recommendations. Genet. Med. J. Am. Coll. Med. Genet. 26, 101213 (2024).
Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 30, 2114–2120 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J. & Peterson, H. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Research 9, ELIXIR-709 (2020).
Nava, C. et al. Dominant variants in major spliceosome U4 and U5 small nuclear RNA genes cause neurodevelopmental disorders through splicing disruption. Nat. Genet. 57, 1374–1388 (2025).
Acknowledgements
The authors wish to thank the patients and families included as part of this study. The authors acknowledge HUGODIMS (Western France exome-based trio approach project to identify genes involved in intellectual disability); funding for HUGODIMS is supported by a grant from the French Ministry of Health and from the Health Regional Agency from Poitou-Charentes (HUGODIMS, 2013, RC14_0107). This study was funded by INSERM, by the French League against cancer (la Ligue contre le Cancer, committees 29 and 35), by the French Biomedecine Agency and by the association Gaetan Saleun. T.B. was funded by the Brittany Region and the Ministère de l’Enseignement Supérieur, S.Ch. and C. D. were funded by the Ministère de l’Enseignement Supérieur de la Recherche et de l’Innovation. This study was supported by the « Priority Research Programme on Rare Diseases » of the French Investments for the Future Programme, project MultiOmixCare. JRL was supported in part by US National Institutes of Health NS105078 and HG011758. D.G.C. was supported by the Child Neurologist Career Development Programme K12 and Muscular Dystrophy Association Development Grant (873841). The authors thank the vectorology core facility Vect’UB in Bordeaux for the production of lentiviral particles, and thank the Centre of Biological Resources in Brest (CHU).
Author information
Authors and Affiliations
Contributions
K.U. and T.B. contributed equally. K.U., T.B., and S.K. contributed to the design of the study; T.B., S.Ch., C.D., S.C. performed the experiments; M.P.S.-B. performed the bioinformatic analysis (RNAseq); D.G.B. and K.U. contributed to the bioinformatic analysis; T.Be., L.D.S.F and B. Co. provided data on human samples; K.U., T.B., S.K., S.Ch. and D.G.B. contributed to data interpretation; M.P., S.H., P.W., H.W., B.X., V.S., M.C., C.Z., C.C., T.P., S.G., B.T., T.C., C.P., T.H., T.R., M.M., K.I., J.L., E.Z., I.G., S.W., M.B., L.F., J.M., J.D., C.J., L.LM., H.VE., D.C., L.DW., G.B., C.Pe. and L.D. contributed to the clinical data collection; K.U., S.K., C.G., M.C., S.B., P.S., Y.X., Y.W., M.D-F., D.L., P.C., F.T-M-T., A.D-P., A.V., J. Lu., P.P-B., R.M. and D.S. contributed to the exome/genome data analysis; C.F. supported the study; K.U., S.K., T.B. and D.B wrote the manuscript; E.L., L.C. reviewed the manuscript; D.G.B. designed and supervised the research. All authors read the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Juan Valcárcel Juarez, Christel Depienne, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Uguen, K., Bergot, T., Scott-Boyer, MP. et al. De novo variants in the splicing factor gene SF3B1 are associated with neurodevelopmental disorders. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68284-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68284-9