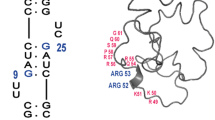
Abstract
Hexim proteins are key RNA-dependent regulators of eukaryotic transcription through 7SK-dependent sequestration and inactivation of the kinase P-TEFb (Cdk9–CyclinT1/2) in the 7SK RNP. P-TEFb activity drives release of RNA polymerase II from promoter-proximal pausing for eukaryotic and HIV-1 transcription. The molecular mechanism by which 7SK binding overcomes an intrinsic Hexim autoinhibition for subsequent P-TEFb inactivation has remained unresolved. Here, using NMR and biophysical methods we demonstrate that Hexim1 homodimer engages two high-affinity sites on 7SK RNA. This dual-site binding triggers a conformational rearrangement in Hexim1’s disordered central region that unmasks the Cdk9-binding site, which is otherwise sequestered within an inter-monomer dimer interface. These findings reveal how Hexim autoinhibition dictates its specificity for 7SK RNA and prevents premature P-TEFb inhibition in the absence of 7SK, thereby providing a mechanistic understanding of Hexim/P-TEFb assembly into the 7SK RNP and further considerations for understanding Hexim–Tat competition during viral transcription.
Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. Raw data analyses are included as a combined supplementary data spreadsheet for protein CSPs, protein PRE intensity ratios, and RNA CSPs. Backbone chemical shift assignments of BR-L-AR protein, and backbone/sidechain chemical shift assignments of SL1-dIΔU RNA-bound BR-L-AR protein have been deposited in the Biological Magnetic Resonance Data Bank (BMRB), under accession IDs 53441 and 53442, respectively. Backbone and sidechain chemical shift assignments of additional monomeric protein constructs BR-L and BR have been deposited in BMRB under accession IDs 53446 and 53447, respectively. Proton chemical shift assignments of RNA constructs, SL1-dI (with additional carbon chemical shifts), SL1-dIΔU, SL1-dII, SL1-d, SL1-dIIm, SL1-p, SL1-mp, SL1-pII and SL1altUUCG, have been deposited in BMRB under accession IDs 53443, 53444, 53445, 53448, 53449, 53450, 53451, 53452, 53453, 53454, respectively. Source data for the figures and Supplementary Figures are provided as a Source Data file. Source data are provided in this paper.
References
Kusuhara, M. et al. Cloning of hexamethylene-bis-acetamide-inducible transcript, HEXIM1, in human vascular smooth muscle cells. Biomed. Res. 20, 273–279 (1999).
Michels, A. A. & Bensaude, O. Hexim1, an RNA-controlled protein hub. Transcription 9, 262–271 (2018).
Morchikh, M. et al. HEXIM1 and NEAT1 Long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immuneresponse. Mol. Cell 67, 387–399 (2017).
Fujimoto, Y., Nakamura, Y. & Ohuchi, S. HEXIM1-binding elements on mRNAs identified through transcriptomic SELEX and computational screening. Biochimie 94, 1900–1909 (2012).
Yik, J. H., Chen, R., Pezda, A. C. & Zhou, Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 280, 16368–16376 (2005).
Byers, S. A., Price, J. P., Cooper, J. J., Li, Q. & Price, D. H. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 280, 16360–16367 (2005).
Michels, A. A. et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23, 2608–2619 (2004).
Yik, J. H., Chen, R., Pezda, A. C., Samford, C. S. & Zhou, Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 24, 5094–5105 (2004).
Czudnochowski, N., Bösken, C. A. & Geyer, M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3, 842 (2012).
Czudnochowski, N., Vollmuth, F., Baumann, S., Vogel-Bachmayr, K. & Geyer, M. Specificity of Hexim1 and Hexim2 complex formation with cyclin T1/T2, importin alpha and 7SK snRNA. J. Mol. Biol. 395, 28–41 (2010).
Graham, T. G. W. et al. Single-molecule live imaging of subunit interactions and exchange within cellular regulatory complexes. Mol. Cell 85, 2854–2868.e2857 (2025).
Marshall, N. F. & Price, D. H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338 (1995).
Vos, S. M. et al. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612 (2018).
D’Orso, I. & Frankel, A. D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 17, 815–821 (2010).
Lu, H. et al. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proc. Natl. Acad. Sci. USA 111, E15–E24 (2014).
D’Orso, I. et al. Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA. Mol. Cell. Biol. 32, 4780–4793 (2012).
Barboric, M. et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35, 2003–2012 (2007).
Dames, S. A. et al. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. USA 104, 14312–14317 (2007).
Schulte, A. et al. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280, 24968–24977 (2005).
Bayer, T. S., Booth, L. N., Knudsen, S. M. & Ellington, A. D. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11, 1848–1857 (2005).
Kobbi, L. et al. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc. Natl. Acad. Sci. USA 113, 12721–12726 (2016).
Barboric, M. et al. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 24, 4291–4303 (2005).
Li, Q. et al. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 35, 2503–2512 (2007).
Lebars, I. et al. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 38, 7749–7763 (2010).
Bourbigot, S. et al. Solution structure of the 5’-terminal hairpin of the 7SK small nuclear RNA. RNA 22, 1844–1858 (2016).
Pham, V. V. et al. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 9, 4266 (2018).
Pham, V. V., Gao, M., Meagher, J. L., Smith, J. L. & D’Souza, V. M. A structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA. Commun. Biol. 5, 819 (2022).
Gruber, A. R. et al. Arthropod 7SK RNA. Mol. Biol. Evol. 25, 1923–1930 (2008).
Gruber, A. R. et al. Invertebrate 7SK snRNAs. J. Mol. Evol. 66, 107–115 (2008).
Muniz, L., Egloff, S., Ughy, B., Jády, B. E. & Kiss, T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 6, e1001152 (2010).
Yang, Y., Eichhorn, C. D., Wang, Y., Cascio, D. & Feigon, J. Structural basis of 7SK RNA 5’-γ-phosphate methylation and retention by MePCE. Nat. Chem. Biol. 15, 132–140 (2019).
Marz, M. et al. Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 26, 2821–2830 (2009).
Brogie, J. E. & Price, D. H. Reconstitution of a functional 7SK snRNP. Nucleic Acids Res. 45, 6864–6880 (2017).
Yang, Y. et al. Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP. Mol. Cell 82, 1724–1736 (2022).
Olson, S. W. et al. Discovery of a large-scale, cell-state-responsive allosteric switch in the 7SK RNA using DANCE-MaP. Mol. Cell 82, 1708–1723 (2022).
Xue, Y., Yang, Z., Chen, R. & Zhou, Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 38, 360–369 (2010).
Van Herreweghe, E. et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26, 3570–3580 (2007).
Schägger, H. & von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 (1991).
Sonn-Segev, A. et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 11, 1772 (2020).
Young, G. et al. Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018).
Bugge, K. et al. Role of charges in a dynamic disordered complex between an IDP and a folded domain. Nat. Commun. 16, 3242 (2025).
Marsh, J. A., Singh, V. K., Jia, Z. & Forman-Kay, J. D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 (2006).
Brillet, K. et al. Different views of the dynamic landscape covered by the 5’-hairpin of the 7SK small nuclear RNA. RNA 26, 1184–1197 (2020).
Martinez-Zapien, D. et al. The crystal structure of the 5′ functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 45, 3568–3579 (2017).
Zhang, Q., Sun, X., Watt, E. D. & Al-Hashimi, H. M. Resolving the motional modes that code for RNA adaptation. Science 311, 653–656 (2006).
Meyer, B. & Peters, T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 42, 864–890 (2003).
Battiste, J. L. & Wagner, G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39, 5355–5365 (2000).
Clore, G. M. & Iwahara, J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 109, 4108–4139 (2009).
Tang, C., Ghirlando, R. & Clore, G. M. Visualization of transient ultra-weak protein self-association in solution using paramagnetic relaxation enhancement. J. Am. Chem. Soc. 130, 4048–4056 (2008).
Morris, O. M., Torpey, J. H. & Isaacson, R. L. Intrinsically disordered proteins: modes of binding with emphasis on disordered domains. Open Biol. 11, 210222 (2021).
AJ, C. Q., Bugai, A. & Barboric, M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 44, 7527–7539 (2016).
Contreras, X., Barboric, M., Lenasi, T. & Peterlin, B. M. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3, 1459–1469 (2007).
Kim, Y. K., Mbonye, U., Hokello, J. & Karn, J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410, 896–916 (2011).
Mbonye, U. R., Wang, B., Gokulrangan, G., Chance, M. R. & Karn, J. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 promotes release of P-TEFb from the 7SK snRNP complex and enhances proviral HIV gene expression. Proteomics 15, 2078–2086 (2015).
Fujinaga, K. et al. PKC phosphorylates HEXIM1 and regulates P-TEFb activity. Nucleic Acids Res. 40, 9160–9170 (2012).
Sun, Y. et al. Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease. Sci. Adv. 5, eaaw3593 (2019).
Egloff, S., Van Herreweghe, E. & Kiss, T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell Biol 26, 630–642 (2006).
Zeke, A. et al. Deep structural insights into RNA-binding disordered protein regions. Wiley Interdiscip. Rev. RNA. 13, e1714 (2022).
Hentze, M. W., Castello, A., Schwarzl, T. & Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018).
Qiu, C. et al. Intra- and inter-molecular regulation by intrinsically-disordered regions governs PUF protein RNA binding. Nat. Commun. 14, 7323 (2023).
Chong, P. A., Vernon, R. M. & Forman-Kay, J. D. RGG/RG Motif Regions in RNA Binding and Phase Separation. J. Mol. Biol. 430, 4650–4665 (2018).
Wang, X. et al. Negatively charged, intrinsically disordered regions can accelerate target search by DNA-binding proteins. Nucleic Acids Res. 51, 4701–4712 (2023).
Eichhorn, C. D., Yang, Y., Repeta, L. & Feigon, J. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc. Natl. Acad. Sci. USA 115, E6457–e6466 (2018).
Li, Q. et al. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280, 28819–28826 (2005).
Prasanth, K. V. et al. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell 21, 4184–4196 (2010).
Wang, Y. et al. N(6)-methyladenosine in 7SK small nuclear RNA underlies RNA polymerase II transcription regulation. Mol. Cell 83, 3818–3834 (2023).
Perez-Pepe, M. et al. 7SK methylation by METTL3 promotes transcriptional activity. Sci. Adv. 9, eade7500 (2023).
Mbonye, U., Kizito, F. & Karn, J. New insights into transcription elongation control of HIV-1 latency and rebound. Trends Immunol. 44, 60–71 (2023).
Sobhian, B. et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 38, 439–451 (2010).
Cheng, Y. & Patel, D. J. An efficient system for small protein expression and refolding. Biochem. Biophys. Res. Commun. 317, 401–405 (2004).
Sjodt, M. & Clubb, R. T. Nitroxide labeling of proteins and the determination of paramagnetic relaxation derived distance restraints for NMR studies. Bio Protoc. 7, https://doi.org/10.21769/BioProtoc.2207 (2017).
Guillerez, J., Lopez, P. J., Proux, F., Launay, H. & Dreyfus, M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. USA 102, 5958–5963 (2005).
Hyberts, S. G., Milbradt, A. G., Wagner, A. B., Arthanari, H. & Wagner, G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327 (2012).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Clamp, M., Cuff, J., Searle, S. M. & Barton, G. J. The Jalview Java alignment editor. Bioinformatics 20, 426–427 (2004).
Acknowledgements
This work was supported by the National Institutes of Health (NIH) R01AI155170 and R35GM131901 to J.F., American Heart Association Postdoctoral Fellowship 20POST35210850 to Y.Y. and American-Italian Cancer Foundation Postdoctoral Fellowship to M.G.M. The UCLA-DOE NMR core facility is supported in part by NIH instrumentation grants S10OD016336 and S10OD025073 and Department of Energy DE-FC02-02ER63421. We thank Catherine D. Eichhorn and Yanjiao Wang for helpful discussions.
Author information
Authors and Affiliations
Contributions
Y.Y., M.G.M., and J.F. conceptualized the study and designed the experiments. Y.Y. and M.G.M. performed the majority of the experiments and data analysis. S.G. prepared RNA and protein samples and acquired NMR, NativePAGE, and mass photometry data. Y.W. designed SL1 proximal constructs and performed NMR assignments. C.S., N.A., and X.W. helped with sample preparation for NMR and ITC experiments. Y.Y. and J.F. wrote the manuscript, and all authors participated in editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Murrali, M.G., Galvan, S. et al. HEXIM1 inter-monomer autoinhibition governs 7SK RNA binding specificity and P-TEFb inactivation. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68285-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68285-8