Abstract
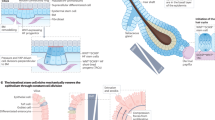
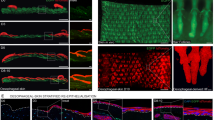
The skin exhibits extraordinary plasticity, enabling it to adapt to mechanical changes in the environment. While transient deformations are accommodated without lasting structural effects, sustained mechanical stress induces durable tissue changes. To investigate if these responses are mediated by shifts in epidermal stem cell fate, we employed two-photon intravital imaging to visualize epidermal cells in live skin subjected to acute mechanical forces. Mechanical force triggered the formation of intracellular “stress” vesicles within epidermal stem cells that filled with extracellular fluid and progressively enlarged, deforming the nucleus. Lineage tracing analyses revealed that the extent of nuclear deformation can predict cell fate outcomes. Moreover, mechanical stress caused sustained elevation of intracellular calcium in basal epidermal stem cells, and conditional deletion of the mechanosensitive ion channel Piezo1 disrupted calcium dynamics and increased stress vesicle formation. Using human skin xenografts, we demonstrated that stress vesicles are conserved in mammalian skin. Together, these findings identify stress vesicles as key mediators linking mechanical stress, calcium signaling, and epidermal stem cell fate.
Similar content being viewed by others
Data availability
Reagents presented in this study are available from the corresponding author upon reasonable request. RNA sequencing data can be accessed through the Gene Expression Omnibus under accession number GSE217491. Source data are provided with this paper.
References
Joodaki, H. & Panzer, M. B. Skin mechanical properties and modeling: a review. Proc. Inst. Mech. Eng. Part H. J. Eng. Med. 232, 323–343 (2018).
Biggs, L. C., Kim, C. S., Miroshnikova, Y. A. & Wickström, S. A. Mechanical forces in the skin: roles in tissue architecture, stability, and function. J. Invest. Dermatol. 140, 284–290 (2019).
Filippo, R. E. D. & Atala, A. Stretch and growth: the molecular and physiologic influences of tissue expansion. Plast. Reconstr. Surg. 109, 2450–2462 (2002).
Tepole, A. B., Gosain, A. K. & Kuhl, E. Stretching skin: the physiological limit and beyond. Int J. Nonlinear Mech. 47, 938–949 (2012).
Yang, W. et al. On the tear resistance of skin. Nat. Commun. 6, 6649 (2015).
Oxlund, H., Manschot, J. & Viidik, A. The role of elastin in the mechanical properties of skin. J. Biomech. 21, 213–218 (1988).
Huang, X. et al. Matrix stiffness–induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am. J. Resp. Cell Mol. 47, 340–348 (2012).
Balaban, N. Q. et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 (2001).
Plikus, M. V. et al. Fibroblasts: origins, definitions, and functions in health and disease. Cell 184, 3852–3872 (2021).
Le, H. Q. et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18, 864–875 (2016).
Carley, E. et al. The LINC complex transmits integrin-dependent tension to the nuclear lamina and represses epidermal differentiation. eLife 10, e58541 (2021).
Lamaze, C. & Torrino, S. Caveolae and cancer: a new mechanical perspective. Biomed. J. 38, 367 (2015).
Swift, J. & Discher, D. E. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 127, 3005–3015 (2014).
Alisafaei, F., Jokhun, D. S., Shivashankar, G. V. & Shenoy, V. B. Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl. Acad. Sci. 116, 13200–13209 (2019).
Pfeifer, C. R., Alvey, C. M., Irianto, J. & Discher, D. E. Genome variation across cancers scales with tissue stiffness – an invasion-mutation mechanism and implications for immune cell infiltration. Curr. Opin. Syst. Biol. 2, 102–113 (2017).
LeGoff, L. & Lecuit, T. Mechanical forces and growth in animal tissues. Csh Perspect. Biol. 8, a019232 (2015).
Matamoro-Vidal, A. & Levayer, R. Multiple influences of mechanical forces on cell competition. Curr. Biol. 29, R762–R774 (2019).
Jaalouk, D. E. & Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Bio 10, 63–73 (2009).
Aureille, J., Belaadi, N. & Guilluy, C. Mechanotransduction via the nuclear envelope: a distant reflection of the cell surface. Curr. Opin. Cell Biol. 44, 59–67 (2017).
Stan, R. V. Structure of caveolae. Biochim. Biophys. Acta Bba - Mol. Cell Res 1746, 334–348 (2005).
Sinha, B. et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144, 402–413 (2011).
Joseph, J. G. & Liu, A. P. Mechanical regulation of endocytosis: new insights and recent advances. Adv. Biosyst. 4, 1900278 (2020).
Gudipaty, S. A. et al. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 543, 118–121 (2017).
Apodaca, G. Modulation of membrane traffic by mechanical stimuli. Am. J. Physiol.-Ren. 282, F179–F190 (2002).
Nava, M. M. et al. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800–817.e22 (2020).
Aragona, M. et al. Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 584, 268–273 (2020).
Khilan, A. A., Al-Maslamani, N. A. & Horn, H. F. Cell stretchers and the LINC complex in mechanotransduction. Arch. Biochem Biophys. 702, 108829 (2021).
Senoo, M. Epidermal stem cells in homeostasis and wound repair of the skin. Adv. Wound Care 2, 273–282 (2013).
Kaur, P. Interfollicular epidermal stem cells: identification, challenges, potential. J. Invest Dermatol 126, 1450–1458 (2006).
Lim et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Sci. N. Y. N. Y 342, 1226–1230 (2013).
Dekoninck, S. & Blanpain, C. Stem cell dynamics, migration and plasticity during wound healing. Nat. Cell Biol. 21, 18–24 (2019).
Martino, F., Perestrelo, A. R., Vinarský, V., Pagliari, S. & Forte, G. Cellular mechanotransduction: from tension to function. Front Physiol. 9, 824 (2018).
Liu, Y.-S. & Lee, O. K. In search of the pivot point of mechanotransduction: mechanosensing of stem cells. Cell Transpl. 23, 1–11 (2012).
Gerard, J., Borst, G. & Helmchen, F. Calcium influx during an action potential. Methods Enzymol. 293, 352–371 (1998). [20].
Cho, C.-H., Woo, J. S., Perez, C. F. & Lee, E. H. A focus on extracellular Ca2+ entry into skeletal muscle. Exp. Mol. Med. 49, e378–e378 (2017).
Snoeck, H. Calcium regulation of stem cells. Embo Rep. 21, e50028 (2020).
Miroshnikova, Y. A. et al. Calcium signaling mediates a biphasic mechanoadaptive response of endothelial cells to cyclic mechanical stretch. Mol. Biol. Cell 32, 1724–1736 (2021).
Passini, F. S. et al. Shear-stress sensing by PIEZO1 regulates tendon stiffness in rodents and influences jumping performance in humans. Nat. Biomed. Eng. 5, 1457–1471 (2021).
Li, J. et al. Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 (2014).
Friedrich, E. E. et al. Endothelial cell Piezo1 mediates pressure-induced lung vascular hyperpermeability via disruption of adherens junctions. Proc. Natl. Acad. Sci. 116, 12980–12985 (2019).
Kraus, D. et al. Intracellular calcium dynamics - sparks of insight. J. Basic Clin. Physiol. Pharm. 11, 331–366 (2000).
Casteels, R. & Droogmans, G. Cell membrane responsiveness and excitation-contraction coupling in smooth muscle. J. Cardiovasc Pharm. 6, S304–S312 (1984).
Ji, G., Barsotti, R. J., Feldman, M. E. & Kotlikoff, M. I. Stretch-induced calcium release in smooth muscle. J. Gen. Physiol. 119, 533–543 (2002).
Retailleau, K. et al. Piezo1 in smooth muscle cells is involved in hypertension-dependent arterial remodeling. Cell Rep. 13, 1161–1171 (2015).
Chen, X. et al. A feedforward mechanism mediated by mechanosensitive ion channel PIEZO1 and tissue mechanics promotes glioma aggression. Neuron 100, 799–815.e7 (2018).
Syeda, R. et al. Piezo1 channels are inherently mechanosensitive. Cell Rep. 17, 1739–1746 (2016).
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010).
Gnanasambandam, R., Bae, C., Gottlieb, P. A. & Sachs, F. Ionic selectivity and permeation properties of human PIEZO1 Channels. PLoS ONE 10, e0125503 (2015).
Wang, S. et al. Endothelial cation channel PIEZO1 controls blood pressure by mediating flow-induced ATP release. J. Clin. Invest. 126, 4527–4536 (2016).
Liu, T. et al. Piezo1-mediated Ca2+ activities regulate brain vascular pathfinding during development. Neuron 108, 180–192.e5 (2020).
Ma, S. et al. A role of PIEZO1 in iron metabolism in mice and humans. Cell 184, 969–982.e13 (2021).
Cahalan, S. M. et al. Piezo1 links mechanical forces to red blood cell volume. Elife 4, e07370 (2015).
Murthy, S. E., Dubin, A. E. & Patapoutian, A. Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Bio 18, 771–783 (2017).
Eid, E. S. & Kurban, M. S. A Piez-o the jigsaw: the Piezo1 channel in skin biology. Clin. Exp. Dermatol 47, 1036–1047 (2022).
Eisenhoffer, G. T. et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549 (2012).
He, L., Si, G., Huang, J., Samuel, A. D. T. & Perrimon, N. Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 555, 103–106 (2018).
Cox, C. D. et al. Removal of the mechanoprotective influence of the cytoskeleton reveals that PIEZO1 is gated by bilayer tension. Nat. Commun. 7, 10366 (2016).
Lewis, A. H. & Grandl, J. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife 4, e12088 (2015).
Versaevel, M., Riaz, M., Grevesse, T. & Gabriele, S. Cell confinement: putting the squeeze on the nucleus. Soft Matter 9, 6665–6676 (2013).
Lele, T. P., Dickinson, R. B. & Gundersen, G. G. Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 217, 3330–3342 (2018).
Kalukula, Y., Stephens, A. D., Lammerding, J. & Gabriele, S. Mechanics and functional consequences of nuclear deformations. Nat. Rev. Mol. Cell Bio. 23, 583–602 (2022).
Jones, M. L. et al. The elephant in the cell: nuclear mechanics and mechanobiology. J. Biomech. Eng. 144, 080802 (2022).
Amiad-Pavlov, D. et al. Live imaging of chromatin distribution reveals novel principles of nuclear architecture and chromatin compartmentalization. Sci. Adv. 7, eabf6251 (2021).
Srivastava, L. K., Ju, Z., Ghagre, A. & Ehrlicher, A. J. Spatial distribution of lamin A/C determines nuclear stiffness and stress-mediated deformation. J. Cell Sci. 134, jcs248559 (2021).
Goelzer, M., Goelzer, J., Ferguson, M. L., Neu, C. P. & Uzer, G. Nuclear envelope mechanobiology: linking the nuclear structure and function. Nucleus 12, 90–114 (2021).
Coué, M., Brenner, S. L., Spector, I. & Korn, E. D. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316–318 (1987).
Liu, Z. et al. Blebbistatin inhibits contraction and accelerates migration in mouse hepatic stellate cells. Brit J. Pharm. 159, 304–315 (2010).
Li, Y. & Burridge, K. Cell-cycle-dependent regulation of cell adhesions: adhering to the schedule. Bioessays 41, 1800165 (2019).
Jones, M. C., Zha, J. & Humphries, M. J. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. B 374, 20180227 (2019).
Moore, J. L. et al. Cell cycle controls long-range calcium signaling in the regenerating epidermis. J. Cell Biol. 222, e202302095 (2023).
Bayarmagnai, B. et al. Invadopodia-mediated ECM degradation is enhanced in the G1 phase of the cell cycle. J. Cell Sci. 132, jcs227116 (2019).
Munaron, L., Antoniotti, S. & Lovisolo, D. Intracellular calcium signals and control of cell proliferation: how many mechanisms?. J. Cell Mol. Med 8, 161–168 (2004).
Rinnerthaler, M., Streubel, M. K., Bischof, J. & Richter, K. Skin aging, gene expression and calcium. Exp. Gerontol. 68, 59–65 (2015).
Feingold, K. R., Schmuth, M. & Elias, P. M. The regulation of permeability barrier homeostasis. J. Invest Dermatol 127, 1574–1576 (2007).
Maeda, T. et al. Conversion of mechanical force into TGF-β-mediated biochemical signals. Curr. Biol. 21, 933–941 (2011).
Kröger, C. et al. Keratins control intercellular adhesion involving PKC-α–mediated desmoplakin phosphorylation. J. Cell Biol. 201, 681–692 (2013).
Jin, Y.-H. et al. Protein kinase C and calmodulin serve as calcium sensors for calcium-stimulated endocytosis at synapses. J. Neurosci. 39, 9478–9490 (2019).
Gonzalez-Molina, J. et al. Extracellular fluid viscosity enhances liver cancer cell mechanosensing and migration. Biomaterials 177, 113–124 (2018).
Choudhury, M. I., Benson, M. A. & Sun, S. X. Trans-epithelial fluid flow and mechanics of epithelial morphogenesis. Semin Cell Dev. Biol. 131, 146–159 (2022).
Dumortier, J. G. et al. Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365, 465–468 (2019).
Mosaliganti, K. R. et al. Size control of the inner ear via hydraulic feedback. Elife 8, e39596 (2019).
Casares, L. et al. Hydraulic fracture during epithelial stretching. Nat. Mater. 14, 343–351 (2015).
Latorre, E. et al. Active superelasticity in three-dimensional epithelia of controlled shape. Nature 563, 203–208 (2018).
Morris, C. E., Wang, J. A. & Markin, V. S. The Invagination of Excess Surface Area by Shrinking Neurons. Biophys. J. 85, 223–235 (2003).
Kennard, A. S., Sathe, M., Labuz, E. C., Prinz, C. K. & Theriot, J. A. Post-injury hydraulic fracturing drives fissure formation in the zebrafish basal epidermal cell layer. Curr. Biol. 33, 2616 (2023).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Humphrey, J. D., Dufresne, E. R. & Schwartz, M. A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Bio 15, 802–812 (2014).
Charras, G. & Yap, A. S. Tensile forces and mechanotransduction at cell–cell junctions. Curr. Biol. 28, R445–R457 (2018).
Kechagia, J. Z., Ivaska, J. & Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Bio 20, 457–473 (2019).
Elias, P. M. et al. Modulations in epidermal calcium regulate the expression of differentiation specific markers. J. Invest. Dermatol. 119, 1128–1136 (2002).
Wang, D., Chadha, G. K., Feygin, A. & Ivanov, A. I. F-actin binding protein, anillin, regulates integrity of intercellular junctions in human epithelial cells. Cell Mol. Life Sci. 72, 3185–3200 (2015).
Holt, J. R. et al. Spatiotemporal dynamics of PIEZO1 localization controls keratinocyte migration during wound healing. Elife 10, e65415 (2021).
Mikesell, A. R. et al. Keratinocyte PIEZO1 modulates cutaneous mechanosensation. Elife 11, e65987 (2022).
Dupont, S. & Wickström, S. A. Mechanical regulation of chromatin and transcription. Nat. Rev. Genet 23, 624–643 (2022).
Biedzinski, S. et al. Microtubules control nuclear shape and gene expression during early stages of hematopoietic differentiation. Embo J. 39, e103957 (2020).
Yoon, S. & Leube, R. E. Keratin intermediate filaments: intermediaries of epithelial cell migration. Essays Biochem 63, 521–533 (2019).
Santos, M., Paramio, J. M., Bravo, A., Ramirez, A. & Jorcano, J. L. The expression of keratin k10 in the basal layer of the epidermis inhibits cell proliferation and prevents skin tumorigenesis. J. Biol. Chem. 277, 19122–19130 (2002).
Ho, M. et al. Update of the keratin gene family: evolution, tissue-specific expression patterns, and relevance to clinical disorders. Hum. Genomics 16, 1 (2022).
Wang, F., Chen, S., Liu, H. B., Parent, C. A. & Coulombe, P. A. Keratin 6 regulates collective keratinocyte migration by altering cell–cell and cell–matrix adhesion. J. Cell Biol. 217, 4314–4330 (2018).
Russell, D., Andrews, P. D., James, J. & Lane, E. B. Mechanical stress induces profound remodelling of keratin filaments and cell junctions in epidermolysis bullosa simplex keratinocytes. J. Cell Sci. 117, 5233–5243 (2004).
Vassar, R., Coulombe, P. A., Degenstein, L., Albers, K. & Fuchs, E. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell 64, 365–380 (1991).
Jacob, J. T. et al. Keratin 17 regulates nuclear morphology and chromatin organization. J. Cell Sci. 133, jcs254094 (2020).
Redmond, C. J. & Coulombe, P. A. Intermediate filaments as effectors of differentiation. Curr. Opin. Cell Biol. 68, 155–162 (2021).
Huang, S. & Rompolas, P. Two-photon microscopy for intracutaneous imaging of stem cell activity in mice. Exp. Dermatol 26, 379–383 (2017).
Huang, S. et al. Lgr6 marks epidermal stem cells with a nerve-dependent role in wound re-epithelialization. Cell Stem Cell 28, 1582–1596.e6 (2021).
Ridky, T. W., Chow, J. M., Wong, D. J. & Khavari, P. A. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med 16, 1450–1455 (2010).
McNeal, A. S. et al. CDKN2B loss promotes progression from benign melanocytic nevus to melanoma. Cancer Discov. 5, 1072–1085 (2015).
Natale, C. A. et al. Activation of G protein-coupled estrogen receptor signaling inhibits melanoma and improves response to immune checkpoint blockade. Elife 7, e31770 (2018).
Doepner, M. et al. Endogenous DOPA inhibits melanoma through suppression of CHRM1 signaling. Sci. Adv. 8, eabn4007 (2022).
Acknowledgments
We thank George Cotsarelis, John Stanley and John Seykora for their invaluable advice that guided this study. We also thank Sara Wickström, Dennis E. Discher and Jean-Leon Maitre for insightful discussions. We are grateful to the University of Pennsylvania Skin Biology and Disease Research-based Center (SBDRC) for analysis of tissue sections (CPAT Core) and support with the establishment of human-engineered skin xenograft (STaR Core). We also acknowledge the support of the Institute for Regenerative Medicine and the entire stem cell community at Penn. S.H. was supported by an IRM postdoctoral fellowship. G.R. was supported by a training grant (T32GM007229) from NIH/NIGMS. P.K. was supported by the American Association for Cancer Research-John and Elizabeth Leonard Family Foundation Basic Cancer Research Fellowship. P.R. was supported by grants from NIH (R01EY036440) and from the American Cancer Society (RSG1803101DCC). Penn SBDRC was supported by a center core NIH/NIAMS grant (P30AR069589) and a shared instrumentation grant from the NIH Office of the Director (S10OD038384).
Author information
Authors and Affiliations
Contributions
S.H. and P.R. conceptualized the study, designed the experiments, and wrote the manuscript. S.H., P.K., A.B., M.M., G.R., and P.R. performed the experiments. J.Z. and B.C.C. assisted with the RNA sequencing and performed the bioinformatic analysis. P.K., M.D., and T.W.R. assisted with the establishment of human-engineered skin xenograft. T.Z. and S.P. assisted with the histological assays. J.D.B. assisted with the mechanobiology assays. All authors discussed results and participated in the manuscript preparation and editing. P.R. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jean-Léon Maître and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, S., Kuri, P., Zou, J. et al. Stress vesicles link epidermal mechanotransduction to stem cell differentiation. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68294-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68294-7