Abstract
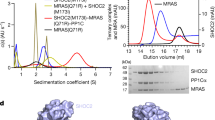
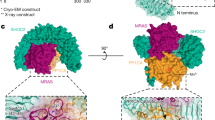
RAF activation is essential for MAPK signaling and is mediated by RAS binding and the dephosphorylation of a conserved phosphoserine by the SHOC2–RAS–PP1C complex. MRAS forms a high-affinity SHOC2–MRAS–PP1C (SMP) complex, while canonical RAS isoforms (KRAS, HRAS, NRAS) form analogous but lower-affinity assemblies. Yet, cancers driven by oncogenic KRAS, HRAS, or NRAS remain strongly SHOC2-dependent, suggesting that these weaker complexes contribute to tumorigenesis. To elucidate how canonical RAS proteins form lower-affinity ternary complexes, the cryo-EM structure of the SHOC2–KRAS–PP1C (SKP) complex stabilized by Noonan syndrome mutations is described. The SKP architecture is similar to the SMP complex but forms fewer contacts and buries less surface area due to the absence of MRAS-specific structural features in KRAS that enhance complex stability. RAS inhibitors MRTX1133 and RMC-6236 alter Switch-I/II conformations, thereby blocking SKP assembly more effectively than they disrupt preformed complexes. These RAS inhibitors do not affect SMP formation because they do not bind MRAS. Since MRAS is upregulated in resistance to KRAS inhibition, we characterize a MRAS mutant capable of binding MRTX1133. This MRAS mutant can form an SMP complex, but MRTX1133 blocks its assembly, demonstrating the feasibility of dual SKP and SMP targeting. Overall, our findings define isoform-specific differences in SHOC2–RAS–PP1C complex formation and support a strategy to prevent both SKP and SMP assemblies to overcome resistance in RAS-driven cancers.
Similar content being viewed by others
Data availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank and can be accessed using accession numbers 9O65, EMD-70159 (stabilized SKP complex), 9O0N (KRAS(1-169)GDP with MRTX1133), 9O0O (KRAS (1-169)GMPPNP with MRTX1133), 9O0P (MRASmut(1-178)GDP with MRTX1133), and 9O0Q (MRASmut(1-178)GMPPNP with MRTX1133). The structures used as initial models for molecular replacement are available in the PDB under accession codes 1X1R (MRAS), 7RPZ (KRAS(G12D)GDP-MRTX1133), and 7TVF (SMP complex). Structures utilized for superpositions, and analysis can be found in the PDB using accession codes 1NVU (HRAS-SOS1), 6OB2 (KRAS + NF1), 6XI7 (KRAS-CRAF), 7LC1 (KRAS-Sin1), 7RPZ (KRAS(G12D)GDP-MRTX1133), 7T47 (KRAS(G12D)GMPPCP-MRTX1133), 7TVF (SMP complex), 8B69 (KRAS-Rgl2), 9AX6 (KRAS-RMC6236-CypA) and 9C15 (KRAS-PI3Kα). The source data underlying Figs. 1d, 5a, 5c, 6a–b, 7b, 7d and 7f–i and Supplementary Figs. 5a, 5g, 6a–b, 7b–c, 8e, 9 and 10 are provided as a Source Data file. Source data are provided with this paper.
References
Lavoie, H. & Therrien, M. Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015).
Prior, I. A., Hood, F. E. & Hartley, J. L. The frequency of RAS mutations in cancer. Cancer Res. 80, 2969–2974 (2020).
Simanshu, D. K., Nissley, D. V. & McCormick, F. RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017).
Freeman, A. K., Ritt, D. A. & Morrison, D. K. The importance of Raf dimerization in cell signaling. Small GTPases 4, 180–185 (2013).
Jeon, H., Tkacik, E. & Eck, M. J. Signaling from RAS to RAF: the molecules and their mechanisms. Annu. Rev. Biochem. 93, 289–316 (2024).
Martinez Fiesco, J. A., Durrant, D. E., Morrison, D. K. & Zhang, P. Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding. Nat. Commun. 13, 486 (2022).
Park, E. et al. Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019).
Mohanty, A. et al. Acquired resistance to KRAS G12C small-molecule inhibitors via genetic/nongenetic mechanisms in lung cancer. Sci. Adv. 9, eade3816 (2023).
Rodriguez-Viciana, P., Sabatier, C. & McCormick, F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell Biol. 24, 4943–4954 (2004).
Ghosh, S. & Bell, R. M. Identification of discrete segments of human Raf-1 kinase critical for high affinity binding to Ha-Ras. J. Biol. Chem. 269, 30785–30788 (1994).
Tran, T. H. et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 12, 1176 (2021).
Young, L. C. et al. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol. Cell 52, 679–692 (2013).
Bonsor, D. A. et al. Structure of the SHOC2-MRAS-PP1C complex provides insights into RAF activation and Noonan syndrome. Nat. Struct. Mol. Biol. 29, 966–977 (2022).
Kwon, J. J. et al. Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex. Nature 609, 408–415 (2022).
Hauseman, Z. J. et al. Structure of the MRAS-SHOC2-PP1C phosphatase complex. Nature 609, 416–423 (2022).
Liau, N. P. D. et al. Structural basis for SHOC2 modulation of RAS signalling. Nature 609, 400–407 (2022).
Snead, K., Wall, V., Ambrose, H., Esposito, D. & Drew, M. Polycistronic baculovirus expression of SUGT1 enables high-yield production of recombinant leucine-rich repeat proteins and protein complexes. Protein Expr. Purif. 193, 106061 (2022).
Yi, J. et al. Endothelial SUR-8 acts in an ERK-independent pathway during atrioventricular cushion development. Dev. Dyn. 239, 2005–2013 (2010).
Nunez Rodriguez, N. et al. Characterization of R-ras3/m-ras null mice reveals a potential role in trophic factor signaling. Mol. Cell Biol. 26, 7145–7154 (2006).
Yi, M., Soppet, D., McCormick, F. & Nissley, D. V. The prominent pervasive oncogenic role and tissue specific permissiveness of RAS gene mutations. Sci. Rep. 14, 25452 (2024).
Bonsor, D. A. & Simanshu, D. K. RAS and SHOC2 roles in RAF activation and therapeutic considerations. Ann. Rev. Cancer Biol. 8, 97–113 (2023).
Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019).
Hallin, J. et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2020).
Adachi, Y. et al. Scribble mis-localization induces adaptive resistance to KRAS G12C inhibitors through feedback activation of MAPK signaling mediated by YAP-induced MRAS. Nat. Cancer 4, 829–843 (2023).
Hagenbeek, T. J. et al. An allosteric pan-TEAD inhibitor blocks oncogenic YAP/TAZ signaling and overcomes KRAS G12C inhibitor resistance. Nat. Cancer 4, 812–828 (2023).
Thatikonda, V. et al. Co-targeting SOS1 enhances the antitumor effects of KRAS(G12C) inhibitors by addressing intrinsic and acquired resistance. Nat. Cancer 5, 1352–1370 (2024).
Zhao, Y. et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 599, 679–683 (2021).
Jiang, J. et al. Translational and therapeutic evaluation of RAS-GTP inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. 14, 994–1017 (2024).
Wang, X. et al. Identification of MRTX1133, a noncovalent, potent, and selective KRAS(G12D) inhibitor. J. Med. Chem. 65, 3123–3133 (2022).
Hallin, J. et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat. Med. 28, 2171–2182 (2022).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Cheung, M. et al. Improved sample dispersion in cryo-EM using “perpetually-hydrated” graphene oxide flakes. J. Struct. Biol. 204, 75–79 (2018).
Bonsor, D. A. & Simanshu, D. K. Structural insights into the role of SHOC2-MRAS-PP1C complex in RAF activation. FEBS J. 290, 4852–4863 (2023).
Peti, W., Nairn, A. C. & Page, R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 280, 596–611 (2013).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Laskowski, R. A., Jablonska, J., Pravda, L., Varekova, R. S. & Thornton, J. M. PDBsum: structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018).
Czyzyk, D. et al. Structural insights into isoform-specific RAS-PI3Kalpha interactions and the role of RAS in PI3Kalpha activation. Nat. Commun. 16, 525 (2025).
Tariq, M. et al. Structural insights into the complex of oncogenic KRas4B(G12V) and Rgl2, a RalA/B activator. Life Sci. Alliance 7, e202302080 (2024).
Castel, P. et al. RAS interaction with Sin1 is dispensable for mTORC2 assembly and activity. Proc. Natl. Acad. Sci. USA 118, e2103261118 (2021).
Yan, W. et al. Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR. Cell Rep. 32, 107909 (2020).
Margarit, S. M. et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003).
Awad, M. M. et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Tanaka, N. & Ebi, H. Mechanisms of resistance to KRAS inhibitors: cancer cells’ strategic use of normal cellular mechanisms to adapt. Cancer Sci. 116, 600–612 (2025).
Rosell, R. et al. KRAS G12C-mutant driven non-small cell lung cancer (NSCLC). Crit. Rev. Oncol. Hematol. 195, 104228 (2024).
Dilly, J. et al. Mechanisms of resistance to oncogenic KRAS inhibition in pancreatic cancer. Cancer Discov. 14, 2135–2161 (2024).
Feng, J. et al. Feedback activation of EGFR/wild-type RAS signaling axis limits KRAS(G12D) inhibitor efficacy in KRAS(G12D)-mutated colorectal cancer. Oncogene 42, 1620–1633 (2023).
Gulay, K. C. M. et al. Dual Inhibition of KRASG12D and pan-ERBB is synergistic in pancreatic ductal adenocarcinoma. Cancer Res. 83, 3001–3012 (2023).
Holderfield, M. et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature 629, 919–926 (2024).
Alexander, P. et al. Biophysical and structural analysis of KRAS switch-II pocket inhibitors reveals allele-specific binding constraints. J. Biol. Chem. 301, 110331 (2025).
Hauseman, Z. J. et al. Targeting the SHOC2-RAS interaction in RAS-mutant cancers. Nature 642, 232–241 (2025).
Jones, G. G. et al. SHOC2 phosphatase-dependent RAF dimerization mediates resistance to MEK inhibition in RAS-mutant cancers. Nat. Commun. 10, 2532 (2019).
Young, L. C. et al. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc. Natl. Acad. Sci. USA 115, 10576–10585 (2018).
Bernal Astrain, G. et al. The small GTPase MRAS is a broken switch. Nat. Commun. 16, 647 (2025).
Ohba, Y. et al. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 275, 20020–20026 (2000).
Quilliam, L. A. et al. M-Ras/R-Ras3, a transforming Ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J. Biol. Chem. 274, 23850–23857 (1999).
Terayama, K. et al. Molecular glues that facilitate RAS binding to PI3Kalpha promote glucose uptake without insulin. Science 389, 402–408 (2025).
Mitin, N., Rossman, K. L. & Der, C. J. Signaling interplay in Ras superfamily function. Curr. Biol. 15, R563–574 (2005).
Young, L. C. & Rodriguez-Viciana, P. MRAS: a close but understudied member of the RAS family. Cold Spring Harb. Perspect. Med. 8, a033621 (2018).
Perurena, N., Situ, L. & Cichowski, K. Combinatorial strategies to target RAS-driven cancers. Nat. Rev. Cancer 24, 316–337 (2024).
Wall, V. E., Garvey, L. A., Mehalko, J. L., Procter, L. V. & Esposito, D. Combinatorial assembly of clone libraries using site-specific recombination. Methods Mol. Biol. 1116, 193–208 (2014).
Grose, C., Wright, C., Mehalko, J. & Esposito, D. Modified E. coli strains enhance baculovirus production by elimination of aberrant transposition events. bioRxiv https://doi.org/10.1101/2021.01.27.427812 (2021).
Talsania, K. et al. Genome assembly and annotation of the Trichoplusia ni Tni-FNL insect cell line enabled by long-read technologies. Genes 10, 79 (2019).
Taylor, T., Denson, J. P. & Esposito, D. Optimizing expression and solubility of proteins in E. coli using modified media and induction parameters. Methods Mol. Biol. 1586, 65–82 (2017).
Han, Y. et al. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 117, 1009–1014 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Lopez-Blanco, J. R. & Chacon, P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 184, 261–270 (2013).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Kabsch, W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Winter, G. et al. DIALS as a toolkit. Protein Sci. 31, 232–250 (2022).
Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. D. Struct. Biol. 79, 449–461 (2023).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 67, 355–367 (2011).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Cuevas-Navarro, A. et al. RAS-dependent RAF-MAPK hyperactivation by pathogenic RIT1 is a therapeutic target in Noonan syndrome-associated cardiac hypertrophy. Sci. Adv. 9, eadf4766 (2023).
Acknowledgements
We acknowledge Dominic Esposito, William Gillette, John-Paul Denson, Matt Drew, Peter H. Frank, Natalie Granato-Guerrero, Brianna Higgins, Min Hong, Jenna Hull, Jennifer Mehalko, Simon Messing, Ashley Mitchell, Shelley Perkins, Ivy Poon, Nitya Ramakrishnan, Katie Geis, Mukul Sherekar, Troy Taylor, and Nicolas Wright for production of protein reagents used in this work. We thank Timothy Waybright for performing the nucleotide exchange analysis. CryoEM data were collected at the NCI National Cryo−EM Facility at the Frederick National Laboratory for Cancer Research, and we thank Tara Fox and Thomas Edwards for their help with the data collection. RASless cells were generated by Matthew J. Sale and Madeleine R. Allison. The authors also acknowledge the use of the Frederick Research Computing Environment (FRCE). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work is based upon research conducted at the NECAT beamlines, which are funded by the NIGMS/NIH (P30 GM124165). This research was supported by an agreement between the Advanced Photon Source and the Diamond Light Source, the U.K.‘s national synchrotron science facility, located at the Harwell Science and Innovation Campus in Oxfordshire, where the work was performed under proposal AU34315-3. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health (NIH) Contract 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
D.A.B. and D.K.S. initiated the project. D.A.B. prepared the cryo-EM sample. L.I.F. and J.R.P. performed negative stain EM, cryo-EM grid preparation, data collection, processing, model building, refinement, and analysis. T.S. produced in-house graphene oxide grids. J.F. helped with the image processing. L.C.Y. and R.G. prepared and performed cellular experiments. D.A.B. conducted crystallography, structural analysis, and ITC experiments. V.E.W. and K.R.G. helped with the cloning and preparation of recombinant proteins. D.A.B., L.C.Y., D.V.N., F.M., and D.K.S. contributed to experimental design and data analysis. D.A.B., L.I.F., and D.K.S. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
F.M. is a consultant for Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Pfizer, Daiichi Sankyo, Amgen, PMV Pharma, OPNA-IO, and Quanta Therapeutics. He has received research grants from Boehringer–Ingelheim, and is a consultant for and cofounder of BridgeBio Pharma. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bonsor, D.A., Finci, L.I., Potter, J.R. et al. Structure of SHOC2-KRAS-PP1C complex reveals RAS isoform-specific determinants and insights into targeting complex assembly by RAS inhibitors. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68319-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68319-1