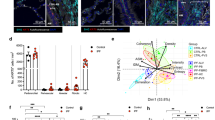
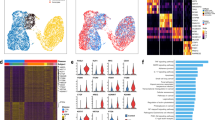
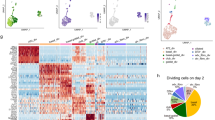
Abstract
Single-cell RNA sequencing (scRNA-seq) has identified intermediate epithelial states in pulmonary fibrosis, including KRT5-/KRT17+ aberrant basaloid cells in humans and Krt8+ alveolar differentiation intermediates (ADIs) in mice. Their functional contributions to fibrogenesis, however, remain unclear. Here, we introduce an RNA-sensing-dependent protein translation technology that enables selective targeting of Krt8+ ADI cells in vitro and in vivo. Transcriptomic analysis revealed Small Proline-Rich Protein 1 A (SPRR1A) mRNA as a shared marker of murine Krt8+ ADIs and human KRT5-/KRT17+ basaloid cells, distinguishing them from other lung cell populations. Using programmable RNA sensors, we demonstrated selective EGFP-labeling of Krt8+ ADI cells in vivo, which faithfully recapitulated their transcriptomic and phenotypic features. To test function, we developed an RNA-sensing-driven diphtheria toxin receptor (DTR) system for conditional ablation of Sprr1a+ cells. Targeted depletion markedly reduced fibrosis in bleomycin-injured mice, establishing transitional epithelial cells as pathogenic drivers and highlighting their potential as therapeutic targets in pulmonary fibrosis.
Similar content being viewed by others
Data availability
The scRNA-seq data generated in this study have been deposited the Gene Expression Omnibus under accession code GSE311198. Source data are provided with this paper.
Code availability
Custom Python scripts used for the single-cell RNA-seq computational analyses have been deposited in a public GitHub repository and archived on Zenodo (https://doi.org/10.5281/zenodo.18027178). The scripts integrate existing open-source libraries to reproduce the analyses described in the previous section; no novel algorithms were developed.
References
Basil, M. C., Alysandratos, K. D., Kotton, D. N. & Morrisey, E. E. Lung repair and regeneration: advanced models and insights into human disease. Cell Stem Cell 31, 439–454 (2024).
El Agha, E. & Thannickal, V. J. The lung mesenchyme in development, regeneration, and fibrosis. J. Clin. Investig. 133, e170498 (2023).
Aspal, M. & Zemans, R. L. Mechanisms of ATII-to-ATI cell differentiation during lung regeneration. Int. J. Mol. Sci. 21, 3188 (2020).
Beers, M. F. & Morrisey, E. E. The three R’s of lung health and disease: repair, remodeling, and regeneration. J. Clin. Investig. 121, 2065–2073 (2011).
Confalonieri, P. et al. Regeneration or repair? The role of alveolar epithelial cells in the pathogenesis of idiopathic pulmonary fibrosis (IPF). Cells 11, 2095 (2022).
Xie, T. et al. Abnormal respiratory progenitors in fibrotic lung injury. Stem Cell Res. Ther. 13, 64 (2022).
Lettieri, S. et al. The plastic interplay between lung regeneration phenomena and fibrotic evolution: current challenges and novel therapeutic perspectives. Int. J. Mol. Sci. 25, 547 (2023).
Strunz, M. et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e367 (2020).
Kobayashi, Y. et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Jiang, P. et al. Ineffectual type 2-to-Type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: persistence of the KRT8. Am. J. Respir. Crit. Care Med. 201, 1443–1447 (2020).
Sun, T. et al. TAZ is required for lung alveolar epithelial cell differentiation after injury. JCI Insight 5, e128674 (2019).
Evans, K. V. & Lee, J. H. Alveolar wars: the rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease. Stem Cells Transl. Med. 9, 867–881 (2020).
Wu, H. et al. Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell 180, 107–121 e117 (2020).
Yao, C. et al. Senescence of alveolar type 2 cells drives progressive pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 203, 707–717 (2021).
Finn, J. et al. Dlk1-mediated temporal regulation of notch signaling is required for differentiation of alveolar type II to type I cells during repair. Cell Rep. 26, 2942–2954.e2945 (2019).
Kathiriya, J. J. et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5. Nat. Cell Biol. 24, 10–23 (2022).
Konkimalla, A. et al. Transitional cell states sculpt tissue topology during lung regeneration. Cell Stem Cell 30, 1486–1502.e1489 (2023).
Wang, F. et al. Regulation of epithelial transitional states in murine and human pulmonary fibrosis. J. Clin. Investig. 133, e165612 (2023).
Qian, Y. et al. Programmable RNA sensing for cell monitoring and manipulation. Nature 610, 713–721 (2022).
Tan, M. H. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Rauch, S. et al. Programmable RNA-guided RNA effector proteins built from human parts. Cell 178, 122–134.e112 (2019).
Limberis, M. P., Vandenberghe, L. H., Zhang, L., Pickles, R. J. & Wilson, J. M. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 17, 294–301 (2009).
Strobel, B. et al. Modeling pulmonary disease pathways using recombinant adeno-associated virus 6.2. Am. J. Respir. Cell Mol. Biol. 53, 291–302 (2015).
van Lieshout, L. P. et al. A novel triple-mutant AAV6 capsid induces rapid and potent transgene expression in the muscle and respiratory tract of mice. Mol. Ther. Methods Clin. Dev. 9, 323–329 (2018).
Kang, M. H. et al. A lung tropic AAV vector improves survival in a mouse model of surfactant B deficiency. Nat. Commun. 11, 3929 (2020).
Kathiriya, J. J., Brumwell, A. N., Jackson, J. R., Tang, X. & Chapman, H. A. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26, 346–358.e344 (2020).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
Sharpless, N. E. & Sherr, C. J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Watanabe, S. et al. Resetting proteostasis with ISRIB promotes epithelial differentiation to attenuate pulmonary fibrosis. Proc. Nat. Acad. Sci. USA 118, e2101100118 (2021).
Ting, C. et al. Biomarkers unveil insights into pathology of transitional epithelial states in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 210, 687–690 (2024).
Reyfman, P. A. et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199, 1517–1536 (2019).
Ting, C. et al. Fatal COVID-19 and non-COVID-19 acute respiratory distress syndrome is associated with incomplete alveolar type 1 epithelial cell differentiation from the transitional state without fibrosis. Am. J. Pathol. 192, 454–467 (2021).
Auyeung, V. C. et al. IRE1alpha drives lung epithelial progenitor dysfunction to establish a niche for pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 322, L564–L580 (2022).
Cervantes-Reyes, A., Smith, A. C., Chinigo, G. M., Blakemore, D. C. & Szostak, M. Decarbonylative Pd-catalyzed suzuki cross-coupling for the synthesis of structurally diverse heterobiaryls. Org. Lett. 24, 1678–1683 (2022).
Murphy, P. Z. & Jester, A. P. Pharmacists’ knowledge and perceptions of health literacy. J. Pharm. Pr. 36, 620–627 (2023).
Prasse, A. et al. BAL cell gene expression is indicative of outcome and airway basal cell involvement in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199, 622–630 (2019).
Selman, M. & Pardo, A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc. Am. Thorac. Soc. 3, 364–372 (2006).
Acknowledgements
The authors would like to thank Dr. Jay K. Kolls and Dr. Kejing Song for their assistance with scRNA-seq. This work was supported in part by NIH grants HL139584, HL156973, and HL174994 (Y.Z.).
Author information
Authors and Affiliations
Contributions
F.P., C.J., Z.Z., and Y.Z. designed the study; F.P., C.J., Z.Z., D.S., A.S., and Y.Q., and C.H. performed experiments; F.P., C.J., Z.Z., S.A., and Y.Z. analyzed data; S.S., C.H., I.O.R., J.A.L., and V.J.T. participated in discussion; F.P., J.S., Z.Z., and Y.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Pinglong Xu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, F., Jiang, Cs., Zheng, Z. et al. Transcriptomic signature-guided depletion of intermediate alveolar epithelial cells ameliorates pulmonary fibrosis in mice. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68354-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68354-y