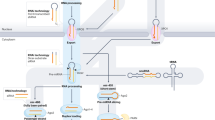
Abstract
MicroRNAs direct Argonaute proteins to repress complementary target mRNAs via mRNA degradation or translational inhibition. While mammalian miRNA targeting has been well studied, the principles by which Drosophila miRNAs bind their target RNAs remain to be fully characterized. Here, we use RNA Bind-n-Seq to systematically identify binding sites and measure their affinities for five highly expressed Drosophila miRNAs. Our results reveal a narrower range of binding site diversity in flies compared to mammals, with fly miRNAs favoring canonical seed-matched sites and exhibiting limited tolerance for imperfections within these sites. We also identified non-canonical site types, including nucleation-bulged and 3′-only sites, whose binding affinities are comparable to canonical sites. These findings establish a foundation for future computational models of Drosophila miRNA targeting, enabling predictions of regulatory outcomes in response to cellular signals, and advancing our understanding of miRNA-mediated regulation in flies.
Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. RBNS sequencing data have been deposited at National Center for Biotechnology Information Sequence Read Archive and are publicly available using accession number PRJNA1185003. The source data underlying Figs. 1c, 2b–c, and 5b, Supplementary Figs. 2b and 5c–d are provided as a Source Data file. Source data are provided with this paper.
Code availability
This study did not generate new code. To estimate KD values, we used the code previously published25 (https://figshare.com/articles/software/MicroRNA-binding_thermodynamics_and_kinetics_by_RNA_Bind-n-Seq/19180952).
References
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Guo, H., Ingolia, N. T., Weissman, J. S. & Bartel, D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Bazzini, A. A., Lee, M. T. & Giraldez, A. J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 (2012).
Doench, J. G. siRNAs can function as miRNAs. Genes Dev. 17, 438–442 (2003).
Doench, J. G. & Sharp, P. A. Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511 (2004).
Hendrickson, D. G. et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7, e1000238 (2009).
Cottrell, K. A., Szczesny, P. & Djuranovic, S. Translation efficiency is a determinant of the magnitude of miRNA-mediated repression. Sci. Rep. 7, 14884 (2017).
Chapat, C. et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. USA 114, 5425–5430 (2017).
Bologna, N. G. & Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in arabidopsis. Annu. Rev. Plant Biol. 65, 473–503 (2014).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Lai, E. C. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30, 363–364 (2002).
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
Stark, A., Brennecke, J., Russell, R. B. & Cohen, S. M. Identification of Drosophila microRNA targets. PLoS Biol. 1, E60 (2003).
Lai, E. C., Tam, B. & Rubin, G. M. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 19, 1067–1080 (2005).
Brennecke, J., Stark, A., Russell, R. B. & Cohen, S. M. Principles of microRNA-target recognition. PLoS Biol. 3, e85 (2005).
Vella, M. C., Choi, E. Y., Lin, S. Y., Reinert, K. & Slack, F. J. T. heC. elegans microRNA let−7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 18, 132–137 (2004).
Grimson, A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 (2007).
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W. & Pasquinelli, A. E. Pairing beyond the seed supports MicroRNA targeting specificity. Mol. Cell 64, 320–333 (2016).
Brancati, G. & Großhans, H. An interplay of miRNA abundance and target site architecture determines miRNA activity and specificity. Nucleic Acids Res. 46, 3259–3269 (2018).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Becker, W. R. et al. High-throughput analysis reveals rules for target RNA binding and cleavage by AGO2. Mol. Cell 75, 741–755.e11 (2019).
McGeary, S. E. et al. The biochemical basis of microRNA targeting efficacy. Science 366, eaav1741 (2019).
Jouravleva, K., Vega-Badillo, J. & Zamore, P. D. Principles and pitfalls of high-throughput analysis of microRNA-binding thermodynamics and kinetics by RNA bind-n-seq. Cell Rep. Methods 2, 100185 (2022).
Shin, C. et al. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38, 789–802 (2010).
Agarwal, V., Subtelny, A. O., Thiru, P., Ulitsky, I. & Bartel, D. P. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 19, 152 (2018).
Kim, D. et al. General rules for functional microRNA targeting. Nat. Genet. 48, 1517–1526 (2016).
Schnall-Levin, M., Zhao, Y., Perrimon, N. & Berger, B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3’UTRs. Proc. Natl. Acad. Sci. USA 107, 15751–15756 (2010).
Lee, R. C., Feinbaum, R. L. & Ambros, V. T. heC. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Hipfner, D. R., Weigmann, K. & Cohen, S. M. The bantam gene regulates Drosophila growth. Genetics 161, 1527–1537 (2002).
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003).
Kadener, S. et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 23, 2179–2191 (2009).
Zhao, C., Sun, G., Li, S. & Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371 (2009).
McGeary, S. E., Bisaria, N., Pham, T. M., Wang, P. Y. & Bartel, D. P. MicroRNA 3’-compensatory pairing occurs through two binding modes, with affinity shaped by nucleotide identity and position. eLife 11, e69803 (2022).
Lambert, N. et al. RNA bind-n-seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 54, 887–900 (2014).
Gainetdinov, I. et al. Relaxed targeting rules help PIWI proteins silence transposons. Nature 619, 394–402 (2023).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Jarmoskaite, I. et al. A quantitative and predictive model for rna binding by human pumilio proteins. Mol. Cell 74, 966–981.e18 (2019).
Zamore, P. D., Bartel, D. P., Lehmann, R. & Williamson, J. R. The PUMILIO−RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38, 596–604 (1999).
Sadée, C. et al. A comprehensive thermodynamic model for RNA binding by the Saccharomyces cerevisiae Pumilio protein PUF4. Nat. Commun. 13, 4522 (2022).
Pasquinelli, A. E. et al. Conservation of the sequence and temporal expression of let−7 heterochronic regulatory RNA. Nature 408, 86–89 (2000).
Sempere, L. F., Dubrovsky, E. B., Dubrovskaya, V. A., Berger, E. M. & Ambros, V. The expression of the let−7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244, 170–179 (2002).
Sokol, N. S., Xu, P., Jan, Y. N. & Ambros, V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22, 1591–1596 (2008).
Chen, W. et al. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat. Commun. 5, 5549 (2014).
Iovino, N., Pane, A. & Gaul, U. miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 17, 123–133 (2009).
Li, P. et al. Localized expression pattern of miR-184 in Drosophila. Mol. Biol. Rep. 38, 355–358 (2011).
Leaman, D. et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121, 1097–1108 (2005).
Weng, R. & Cohen, S. M. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development 139, 1427–1434 (2012).
Garaulet, D. L. et al. miR-124 regulates diverse aspects of rhythmic behavior in drosophila. J. Neurosci. 36, 3414–3421 (2016).
Wee, L. M., Flores-Jasso, C. F., Salomon, W. E. & Zamore, P. D. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151, 1055–1067 (2012).
Schirle, N. T., Sheu-Gruttadauria, J. & MacRae, I. J. Structural basis for microRNA targeting. Science 346, 608–613 (2014).
Salomon, W. E., Jolly, S. M., Moore, M. J., Zamore, P. D. & Serebrov, V. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95 (2015).
Sheu-Gruttadauria, J., Xiao, Y., Gebert, L. F. & MacRae, I. J. Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 38, e101153 (2019).
Chi, S. W., Hannon, G. J. & Darnell, R. B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 19, 321–327 (2012).
Zykovich, A., Korf, I. & Segal, D. J. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 37, e151 (2009).
Okamura, K., Ishizuka, A., Siomi, H. & Siomi, M. C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666 (2004).
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M. C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 (2005).
Förstemann, K., Horwich, M. D., Wee, L., Tomari, Y. & Zamore, P. D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130, 287–297 (2007).
Grubbs, R. D. Intracellular magnesium and magnesium buffering. Biometals 15, 251–259 (2002).
Nielsen, C. B. et al. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13, 1894–1910 (2007).
Tafer, H. et al. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 26, 578–583 (2008).
Wessels, H. H. et al. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat. Commun. 10, 1626 (2019).
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Flores-Jasso, C. F., Salomon, W. E. & Zamore, P. D. Rapid and specific purification of Argonaute-small RNA complexes from crude cell lysates. RNA 19, 271–279 (2013).
Haley, B. In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30, 330–336 (2003).
Pall, G. S. & Hamilton, A. J. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3, 1077–1084 (2008).
Rambo, R. P. & Doudna, J. A. Assembly of an active group II intron− maturase complex by protein dimerization. Biochemistry 43, 6486–6497 (2004).
Lin, S. -y & Riggs, A. D. Lac represser binding to non-operator DNA: detailed studies and a comparison of equilibrium and rate competition methods. J. Mol. Biol. 72, 671–690 (1972).
Weeks, K. M. & Crothers, D. M. RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry 31, 10281–10287 (1992).
Vega-Badillo, J., Zamore, P. D. & Jouravleva, K. Protocol to measure protein-RNA binding using double filter-binding assays followed by phosphorimaging or high-throughput sequencing. STAR Protoc. 4, 102336 (2023).
Smolarsky, M. & Tal, M. Novel method for measuring polyuridylic acid binding to ribosomes. Biochim. Biophys. Acta 199, 447–452 (1970).
Wong, I. & Lohman, T. M. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA 90, 5428–5432 (1993).
Hall, M. H., Wang, P. Y., Pham, T. M. & Bartel, D. P. Functional microRNA targeting without seed pairing. Nucleic Acids Res. 53, gkaf1018 (2025).
Acknowledgements
We thank members of the Zamore and Jouravleva laboratories for critical comments on the manuscript. We thank the KYOTO Drosophila Stock Center (Drosophila Genomics and Genetic Resources, Kyoto Institute of Technology) for providing fly stocks used in this study. This work was supported in part by National Institutes of Health grant R35 GM136275 to P.D.Z. and by ATIP-Avenir funding (CNRS Biology) to K.J.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.V.B, K.J., and P.D.Z.; Methodology, J.V.B. and K.J.; Investigation, J.V.B. and K.J.; Formal analysis, K.J. and J.V.B.; Writing—original draft, K.J. and J.V.B.; Writing—review and editing, K.J., J.V.B., and P.D.Z.; Supervision, K.J. and P.D.Z.; Funding acquisition, K.J. and P.D.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vega-Badillo, J., Zamore, P.D. & Jouravleva, K. Biochemical principles of miRNA targeting in flies. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68360-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68360-0