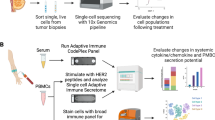
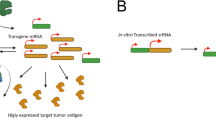
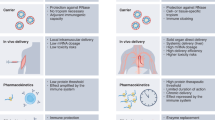
Abstract
Self-amplifying mRNA (saRNA) vectors hold promise for the sustained expression of mRNA vaccines in vivo. However, their inherently high immunogenicity and low-fidelity replication—stemming from the RNA viral genome’s replication mechanisms—limit their efficacy as replacements or adjuncts to protein therapies. Here we report an engineered viral protein genome-linked (VPg) saRNA vector derived from a Norovirus replicon, designed for rapid loading of therapeutic protein mRNAs in vitro. The engineered VPg saRNA is adapted for a range of therapeutic scenarios, including treatment of tumor-associated cachexia under conditions of translational restriction in cap-dependent metabolism, precise encoding of oncolytic mRNAs in vivo to achieve complex functionality, and therapy for graft-versus-host disease in highly auto-immune environments. VPg saRNA addresses key limitations of linear mRNA and conventional saRNA therapies, broadening the potential applications of mRNA-based treatments.
Similar content being viewed by others
Data availability
Source data for Figs. 1–7 and Supplementary Figs. 1–7 are provided as Source Data files. LC–MS/MS raw datasets and Sanger sequencing chromatograms generated in this study have been deposited in Figshare and are available at https://doi.org/10.6084/m9.figshare.3078216546 and https://doi.org/10.6084/m9.figshare.3069415146. Raw fluorescence microscopy, cryo-transmission electron microscopy (cryo-TEM), immunohistochemistry (IHC), and hematoxylin and eosin (H&E) staining images are not publicly available due to large file sizes, instrument-specific formats, and institutional data management restrictions; however, all processed and representative images supporting the findings of this study are included in the paper and its supplementary materials. All other data, including raw imaging data, are available from the corresponding author upon request. Source Data are provided with this paper.
References
Skowronski, D. M. & De Serres, G. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 384, 1576–1577 (2021).
Qiu, X. et al. Development of mRNA vaccines against respiratory syncytial virus (RSV). Cytokine Growth Factor Rev. 68, 37–53 (2022).
Liu, C. et al. mRNA-based cancer therapeutics. Nat. Rev. Cancer 23, 526–543 (2023).
Kim, Y. K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 54, 455–465 (2022).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Anderson, B. R. et al. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 39, 9329–9338 (2011).
Blakney, A. K., Ip, S. & Geall, A. J. An update on self-amplifying mRNA vaccine development. Vaccines 9, 97 (2021).
Rohner, E., Yang, R., Foo, K. S., Goedel, A. & Chien, K. R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 40, 1586–1600 (2022).
Minnaert, A. K. et al. Strategies for controlling the innate immune activity of conventional and self-amplifying mRNA therapeutics: getting the message across. Adv. Drug Deliv. Rev. 176, 113900 (2021).
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023).
Perkovic, M. et al. A trans-amplifying RNA simplified to essential elements is highly replicative and robustly immunogenic in mice. Mol. Ther. 32, 257–259 (2024).
Bloom, K., van den Berg, F. & Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 28, 117–129 (2021).
Schmidt, C. & Schnierle, B. S. Self-amplifying RNA vaccine candidates: alternative platforms for mRNA vaccine development. Pathogens 12, 138 (2023).
Ventoso, I. et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 20, 87–100 (2006).
García, M. A. et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032–1060 (2006).
Rudd, P. A. et al. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J. Virol. 86, 9888–9898 (2012).
Wauquier, N. et al. The acute phase of Chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J. Infect. Dis. 204, 115–123 (2011).
McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Pindel, A. & Sadler, A. The role of protein kinase R in the interferon response. J. Interferon Cytokine Res. 31, 59–70 (2011).
Castro, C., Arnold, J. J. & Cameron, C. E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 107, 141–149 (2005).
Patterson, E. I. et al. Measuring alphavirus fidelity using non-infectious virus particles. Viruses 12, 546 (2020).
Thorne, L. G. & Goodfellow, I. G. Norovirus gene expression and replication. J. Gen. Virol. 95, 278–291 (2014).
Doerflinger, S. Y. et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 13, e1006705 (2017).
Wolff, G., Melia, C. E., Snijder, E. J. & Bárcena, M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 28, 1022–1033 (2020).
Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature 413, 732–738 (2001).
Leen, E. N. et al. A conserved interaction between a C-terminal motif in norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLoS Pathog. 12, e1005379 (2016).
Belliot, G., Sosnovtsev, S. V., Chang, K. O., McPhie, P. & Green, K. Y. Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology 374, 33–49 (2008).
Goodfellow, I. The genome-linked protein VPg of vertebrate viruses-a multifaceted protein. Curr. Opin. Virol. 1, 355–362 (2011).
Högbom, M., Jäger, K., Robel, I., Unge, T. & Rohayem, J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 90, 281–291 (2009).
Yang, X. et al. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527 (2012).
Gong, P. & Peersen, O. B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 107, 22505–22510 (2010).
Liu, W., Shi, X. & Gong, P. A unique intra-molecular fidelity-modulating mechanism identified in a viral RNA-dependent RNA polymerase. Nucleic Acids Res. 46, 10840–10854 (2018).
Rohayem, J., Robel, I., Jäger, K., Scheffler, U. & Rudolph, W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 80, 7060–7069 (2006).
Subba-Reddy, C. V., Goodfellow, I. & Kao, C. C. VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay. J. Virol. 85, 13027–13037 (2011).
Paul, A. V. & Wimmer, E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 206, 12–26 (2015).
te Velthuis, A. J. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 71, 4403–4420 (2014).
Chung, L. et al. Norovirus translation requires an interaction between the C terminus of the genome-linked viral protein VPg and eukaryotic translation initiation factor 4G. J. Biol. Chem. 289, 21738–21750 (2014).
Qin, X., Jiang, B. & Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 15, 781–786 (2016).
Geremia, A. et al. Activation of Akt-mTORC1 signalling reverts cancer-dependent muscle wasting. J. Cachexia Sarcopenia Muscle 13, 648–661 (2022).
Baracos, V. E., Martin, L., Korc, M., Guttridge, D. C. & Fearon, K. C. H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 4, 17105 (2018).
Currow, D. C. & Abernethy, A. P. Anamorelin hydrochloride in the treatment of cancer anorexia-cachexia syndrome. Future Oncol. 10, 789–802 (2014).
Dalton, J. T., Taylor, R. P., Mohler, M. L. & Steiner, M. S. Selective androgen receptor modulators for the prevention and treatment of muscle wasting associated with cancer. Curr. Opin. Support. Palliat. Care 7, 345–351 (2013).
Renz, B. W. et al. β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell 33, 75–90(2018).
Kojima, M. et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660 (1999).
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2+/PTBP1+ pan-adenocarcinoma. Nat. Cancer 5, 30–46 (2024).
Feng, Z. Engineered VPg saRNA achieves cap-independent, low-immunogenic and precise encoding of therapeutic proteins in vivo. figshare https://doi.org/10.6084/m9.figshare.30694151.v1 (2025).
Yan, L. et al. Targeting glucose metabolism sensitizes pancreatic cancer to MEK inhibition. Cancer Res. 81, 4054–4065 (2021).
Griffin, J. H., Zlokovic, B. V. & Mosnier, L. O. Activated protein C: biased for translation. Blood 125, 2898–2907 (2015).
Ranjan, S. et al. Activated protein C protects from GvHD via PAR2/PAR3 signalling in regulatory T-cells. Nat. Commun. 8, 311 (2017).
Sinha, R. K. et al. Activated protein C ameliorates chronic graft-versus-host disease by PAR1-dependent biased cell signaling on T cells. Blood 134, 776–781 (2019).
Sanjuán, R., Nebot, M. R., Chirico, N., Mansky, L. M. & Belshaw, R. Viral mutation rates. J Virol 84, 9733–9748 (2010).
Poirier, E. Z. et al. Low-fidelity polymerases of alphaviruses recombine at higher rates to overproduce defective interfering particles. J. Virol. 90, 2446–2454 (2015).
Langsjoen, R. M., Muruato, A. E., Kunkel, S. R., Jaworski, E. & Routh, A. Differential alphavirus defective RNA diversity between intracellular and extracellular compartments is driven by subgenomic recombination events. mBio 11, e00731–20 (2020).
Anderson, B. R. et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010).
Kormann, M. S. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011).
McGee, J. E. et al. Complete substitution with modified nucleotides in self-amplifying RNA suppresses the interferon response and increases potency. Nat. Biotechnol. 43, 720–726 (2025).
Lei, J. & Hilgenfeld, R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 591, 3190–3210 (2017).
Koudelka, T. et al. N-terminomics for the identification of in vitro substrates and cleavage site specificity of the SARS-CoV-2 main protease. Proteomics 21, e2000246 (2021).
Chhabra, P. et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 100, 1393–1406 (2019).
Warzak, D. A., Pike, W. A. & Luttgeharm, K. D. Capillary electrophoresis methods for determining the IVT mRNA critical quality attributes of size and purity. SLAS Technol. 28, 369–374 (2023).
Cooke, K. R. et al. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood 88, 3230–3239 (1996).
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32471002, 82401829, 82472617, and 82404553; F.Z.Y.), the National University of Singapore (NUHSRO/2020/133/Startup/08, NUHSRO/2023/008/NUSMed/TCE/LOA, NUHSRO/2021/034/TRP/09/Nanomedicine, NUHSRO/2021/044/Kickstart/09/LOA, 23-0173-A0001; C.X.Y.), the National Medical Research Council (MOH-001388-00, CG21APR1005, MOH-001500-00, MOH-001609-00; C.X.Y.), the Singapore Ministry of Education (MOE-000387-00; C.X.Y.), the National Research Foundation (NRF-000352-00; C.X.Y.), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2023R01002; Z.G.W.), the Distinguished Young Scientists Fund of Zhejiang (LR25H250001; Z.G.W.), and the National Science and Technology Major Project of China (No. 2025ZD1802201; Z.G.W.).
Author information
Authors and Affiliations
Contributions
Z.Y.F. conceived and designed the overall study, supervised key experiments, and led data analysis and interpretation. Z.Y.F. and L.X.C. performed most molecular and cellular experiments. J.Z. and P.W. assisted with cell culture and animal studies. Q.L., Z.L.X., L.Y., and Y.J.H. contributed to data processing, statistical analysis, and figure preparation. X.B.Z., J.H.Z., and Q.C. provided essential reagents, technical guidance, and methodological support. Y.B.P. performed imaging and histopathological examinations. X.K.L., Z.G.W., and X.Y.C. jointly supervised the project, contributed to conceptual refinement, and provided funding and resources. Z.Y.F. wrote the manuscript with input from all authors. All authors discussed the results, revised the manuscript, and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
Xiaoyuan Chen is a co-founder of and holds shares in Yantai Lannacheng Biotechnology Co., Ltd. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, Z., Chu, L., Li, Q. et al. Engineered VPg saRNA achieves cap-independent, low-immunogenic and precise encoding of therapeutic proteins in vivo. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68364-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68364-w