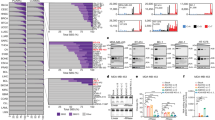
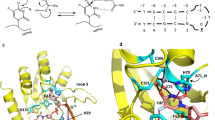
Abstract
APOBEC family members play crucial roles in antiviral restriction. However, certain APOBEC3 (A3) proteins drive harmful hypermutation in humans, contributing to cancer. The cancer-associated A3 proteins are capable of transiting from the cytosol to the nucleus, where they can cause genome mutations. Here, we uncover a specific set of cellular pathways that protect genomic DNA from the major cancer-associated A3 proteins. Through genetic and proteomic screening, we identify UBR4, UBR5, and HUWE1 as key ubiquitin E3 ligases marking cancer-associated A3B and A3H-I for degradation, thereby limiting A3-driven hypermutation. Mechanistically, UBR5 and HUWE1 recognize A3s in the absence of their RNA binding partner, thus promoting proteasomal degradation of APOBEC3 protein that is not engaged in its antiviral cellular function. Depletion or mutation of the E3 ligases in cells and human cancer samples increases A3-driven genome mutagenesis. Our findings reveal that UBR4, UBR5, and HUWE1 are crucial factors in a ubiquitination cascade that maintains human genome stability.
Similar content being viewed by others
Data availability
Cancer genetic analysis data are available from the ICGC Data portal and through granted access of the TCGA Research Network (https://www.cancer.gov/ccg/research/genome-sequencing/tcga). The mass-spectrometry data generated in this study are available in Supplementary Data 2 and have been deposited to the ProteomeXchange consortium via the PRIDE partner repository with the accession code PXD051267
Public cancer whole-genome sequencing data from ICGC are available through the ICGC Data Portal (public) (https://dcc.icgc.org/releases/PCAWG). The NCBI dbGaP data are under restricted access as per NIH policy, access can be obtained through (https://dbgap.ncbi.nlm.nih.gov/home/).The genetic screen data generated in this study are provided as Supplementary Data 1 and in the Source Data file. Source data are provided with this paper.
Code availability
Data and code pertaining to cancer genome analysis and sequence context analysis can be found on GitHub (https://github.com/menchelab/apobex)154.
References
Refsland, E. W. & Harris, R. S. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 371, 1–27 (2013).
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002).
Hultquist, J. F. et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 85, 11220–11234 (2011).
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003).
Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (2003).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Zhang, H. et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98 (2003).
Rogozin, I. B. et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 8, 647–656 (2007).
Conticello, S. G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229 (2008).
Wang, X. et al. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 85, 3142–3152 (2011).
Harari, A., Ooms, M., Mulder, L. C. F. & Simon, V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83, 295–303 (2009).
OhAinle, M., Kerns, J. A., Li, M. M. H., Malik, H. S. & Emerman, M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (2008).
Ooms, M., Majdak, S., Seibert, C. W., Harari, A. & Simon, V. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J. Virol. 84, 7961–7969 (2010).
Li, M. M. H. & Emerman, M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J. Virol. 85, 8197–8207 (2011).
DeWeerd, R. A. et al. Prospectively defined patterns of APOBEC3A mutagenesis are prevalent in human cancers. Cell Rep. 38, 110555 (2022).
Law, E. K. et al. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo. J. Exp. Med 217, e20200261 (2020).
Petljak, M. et al. Mechanisms of APOBEC3 mutagenesis in human cancer cells. Nature 607, 799–807 (2022).
Starrett, G. J. et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 7, 12918 (2016).
Henderson, S. & Fenton, T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol. Med. 21, 274–284 (2015).
Jalili, P. et al. Quantification of ongoing APOBEC3A activity in tumor cells by monitoring RNA editing at hotspots. Nat. Commun. 11, 2971 (2020).
Petljak, M. et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Morganella, S. et al. The topography of mutational processes in breast cancer genomes. Nat. Commun. 7, 11383 (2016).
Carpenter, M. A. et al. Mutational impact of APOBEC3A and APOBEC3B in a human cell line and comparisons to breast cancer. PLoS Genet. 19, e1011043 (2023).
Bergstrom, E. N. et al. Mapping clustered mutations in cancer reveals APOBEC3 mutagenesis of ecDNA. Nature 602, 510–517 (2022).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Dananberg, A., Striepen, J., Rozowsky, J. S. & Petljak, M. APOBEC mutagenesis in cancer development and susceptibility. Cancers 16, 374 (2024).
Hix, M. A., Wong, L., Flath, B., Chelico, L. & Cisneros, G. A. Single-nucleotide polymorphism of the DNA cytosine deaminase APOBEC3H haplotype I leads to enzyme destabilization and correlates with lung cancer. NAR Cancer 2, zcaa023 (2020).
Sanchez, A. et al. Mesoscale DNA features impact APOBEC3A and APOBEC3B deaminase activity and shape tumor mutational landscapes. Nat. Commun. 15, 2370 (2024).
Burns, M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013).
Burns, M. B., Temiz, N. A. & Harris, R. S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 (2013).
Apolonia, L. et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 11, e1004609 (2015).
York, A., Kutluay, S. B., Errando, M. & Bieniasz, P. D. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog. 12, e1005833 (2016).
Iwatani, Y., Takeuchi, H., Strebel, K. & Levin, J. G. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 80, 5992–6002 (2006).
Yang, H., Kim, K., Li, S., Pacheco, J. & Chen, X. S. Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G. Nat. Commun. 13, 7498 (2022).
Dang, Y. et al. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283, 11606–11614 (2008).
Shaban, N. M. et al. The antiviral and cancer genomic DNA deaminase APOBEC3H Is regulated by an RNA-mediated dimerization mechanism. Mol. Cell 69, 75–86.e9 (2018).
Refsland, E. W. et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 10, e1004761 (2014).
Lackey, L. et al. APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 419, 301–314 (2012).
Salamango, D. J. et al. APOBEC3H subcellular localization determinants define zipcode for targeting HIV-1 for restriction. Mol. Cell Biol. 38, e00356-18 (2018).
Chesarino, N. M. & Emerman, M. Polymorphisms in human APOBEC3H differentially regulate ubiquitination and antiviral activity. Viruses 12, 378 (2020).
de Almeida, M. et al. AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature 599, 491–496 (2021).
Vunjak, M. et al. SPOP targets the immune transcription factor IRF1 for proteasomal degradation (Cell Biology) https://doi.org/10.1101/2022.10.10.511567 (2022).
Scinicariello, S. et al. HUWE1 controls tristetraprolin proteasomal degradation by regulating its phosphorylation. Elife 12, e83159 (2023).
Chan, K. et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 47, 1067–1072 (2015).
Jang, G. M. et al. Protein interaction map of APOBEC3 enzyme family reveals deamination-independent role in cellular function. Mol. Cell Proteom. 100755. https://doi.org/10.1016/j.mcpro.2024.100755 (2024).
Gallois-Montbrun, S. et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81, 2165–2178 (2007).
Izumi, T. et al. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J. Virol. 87, 11047–11062 (2013).
Phalora, P. K., Sherer, N. M., Wolinsky, S. M., Swanson, C. M. & Malim, M. H. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J. Virol. 86, 11712–11724 (2012).
Kozak, S. L., Marin, M., Rose, K. M., Bystrom, C. & Kabat, D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281, 29105–29119 (2006).
Ito, F. et al. Understanding the structure, multimerization, subcellular localization and mc selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci. Rep. 8, 3763 (2018).
Zhen, A., Du, J., Zhou, X., Xiong, Y. & Yu, X.-F. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS ONE 7, e38771 (2012).
Polevoda, B. et al. DNA mutagenic activity and capacity for HIV-1 restriction of the cytidine deaminase APOBEC3G depend on whether DNA or RNA binds to tyrosine 315. J. Biol. Chem. 292, 8642–8656 (2017).
Xiao, X., Li, S.-X., Yang, H. & Chen, X. S. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat. Commun. 7, 12193 (2016).
Huthoff, H. & Malim, M. H. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 81, 3807–3815 (2007).
Huthoff, H., Autore, F., Gallois-Montbrun, S., Fraternali, F. & Malim, M. H. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5, e1000330 (2009).
Matsuoka, T. et al. Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA. Nucleic Acids Res. 46, 10368–10379 (2018).
Bohn, J. A. et al. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat. Commun. 8, 1021 (2017).
Grabarczyk, D. B. et al. Architecture of the UBR4 complex, a giant E4 ligase central to eukaryotic protein quality control. Science 389, 909–914 (2025).
Yang, Z. et al. Molecular basis of SIFI activity in the integrated stress response. Nature 643, 1117–1126 (2025).
Shulkina, A. et al. TRIM52 maintains cellular fitness and is under tight proteolytic control by multiple giant E3 ligases. Nat. Commun. 16, 3894 (2025).
Carrillo Roas, S. et al. Convergence of orphan quality control pathways at a ubiquitin chain-elongating ligase. Mol. Cell 85, 815–828.e10 (2025).
Yau, R. G. et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 918–933.e20 (2017).
Perner, J. et al. The mutREAD method detects mutational signatures from low quantities of cancer DNA. Nat. Commun. 11, 3166 (2020).
Cipolla, L. et al. UBR5 interacts with the replication fork and protects DNA replication from DNA polymerase η toxicity. Nucleic Acids Res. 47, 11268–11283 (2019).
Gudjonsson, T. et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 (2012).
Zhang, T., Cronshaw, J., Kanu, N., Snijders, A. P. & Behrens, A. UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc. Natl. Acad. Sci. USA 111, 12091–12096 (2014).
de Vivo, A. et al. The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res 47, 729–746 (2019).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, Aaltonen. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Butler, K. & Banday, A. R. APOBEC3-mediated mutagenesis in cancer: causes, clinical significance and therapeutic potential. J. Hematol. Oncol. 16, 31 (2023).
Durfee, C. et al. Human APOBEC3B promotes tumor development in vivo including signature mutations and metastases. Cell Rep. Med. 4, 101211 (2023).
McCann, J. L. et al. APOBEC3B regulates R-loops and promotes transcription-associated mutagenesis in cancer. Nat. Genet. 55, 1721–1734 (2023).
Middlebrooks, C. D. et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet. 48, 1330–1338 (2016).
Law, E. K. et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016).
Stopak, K. S., Chiu, Y.-L., Kropp, J., Grant, R. M. & Greene, W. C. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 282, 3539–3546 (2007).
Kreisberg, J. F., Yonemoto, W. & Greene, W. C. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203, 865–870 (2006).
Soros, V. B., Yonemoto, W. & Greene, W. C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 3, e15 (2007).
Friew, Y. N., Boyko, V., Hu, W.-S. & Pathak, V. K. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology 6, 56 (2009).
Khan, M. A. et al. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology 4, 48 (2007).
Wittkopp, C. J., Adolph, M. B., Wu, L. I., Chelico, L. & Emerman, M. A single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 12, e1005865 (2016).
Monda, J. K. et al. HAPSTR1 localizes HUWE1 to the nucleus to limit stress signaling pathways. Cell Reports 42 https://doi.org/10.1016/j.celrep.2023.112496 (2023).
Xu, Y., Anderson, D. E. & Ye, Y. The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2, 1–16 (2016).
Tripathi, S. et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 (2015).
Kim, S. T. et al. The N-recognin UBR4 of the N-end rule pathway is required for neurogenesis and homeostasis of cell surface proteins. PLoS ONE 13, e0202260 (2018).
Chen, L. J. et al. HUWE1 plays important role in mouse preimplantation embryo development and the dysregulation is associated with poor embryo development in humans. Sci. Rep. 6, 37928 (2016).
Shearer, R. F. et al. The E3 ubiquitin ligase UBR5 regulates centriolar satellite stability and primary cilia. MBoC 29, 1542–1554 (2018).
Sanchez, A. et al. BMI1–UBR5 axis regulates transcriptional repression at damaged chromatin. Proc. Natl. Acad. Sci. USA 113, 11243–11248 (2016).
Xiang, G. et al. UBR5 targets tumor suppressor CDC73 proteolytically to promote aggressive breast cancer. Cell Death Dis. 13, 1–14 (2022).
Esnault, C., Millet, J., Schwartz, O. & Heidmann, T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 34, 1522–1531 (2006).
Nakaya, Y., Stavrou, S., Blouch, K., Tattersall, P. & Ross, S. R. In vivo examination of mouse APOBEC3- and human APOBEC3A- and APOBEC3G-mediated restriction of parvovirus and herpesvirus infection in mouse models. J. Virol. 90, 8005–8012 (2016).
Moraes, S. N. et al. Evidence linking APOBEC3B genesis and evolution of innate immune antagonism by gamma-herpesvirus ribonucleotide reductases. Elife 11, e83893 (2022).
Cheng, A. Z. et al. Epstein-barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 4, 78–88 (2019).
Zhang, Z. et al. Stably expressed APOBEC3H forms a barrier for cross-species transmission of simian immunodeficiency virus of chimpanzee to humans. PLoS Pathog. 13, e1006746 (2017).
Chen, Y. et al. APOBEC3B edits HBV DNA and inhibits HBV replication during reverse transcription. Antivir. Res. 149, 16–25 (2018).
Bandarra, S. et al. APOBEC3B potently restricts HIV-2 but Not HIV-1 in a Vif-dependent manner. J. Virol. 95, e0117021 (2021).
Mark, K. G. et al. Orphan quality control shapes network dynamics and gene expression. Cell 186, 3460–3475.e23 (2023).
Tsai, J. M. et al. UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability. Mol. Cell 83, 2753–2767.e10 (2023).
Kaisari, S. et al. Role of ubiquitin-protein ligase UBR5 in the disassembly of mitotic checkpoint complexes. Proc. Natl. Acad. Sci. USA 119, e2121478119 (2022).
Hehl, L. A. et al. Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5. Nat. Chem. Biol. 20, 190–200 (2024).
Hodáková, Z. et al. Cryo-EM structure of the chain-elongating E3 ubiquitin ligase UBR5. EMBO J. 42, e113348 (2023).
Wang, F. et al. Structure of the human UBR5 E3 ubiquitin ligase. Structure 31, 541–552.e4 (2023).
Heidelberger, J. B. et al. Proteomic profiling of VCP substrates links VCP to K6-linked ubiquitylation and c-Myc function. EMBO Rep. 19, e44754 (2018).
Hong, J. H. et al. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol. Cell Proteom. 14, 674–685 (2015).
Haakonsen, D. L. et al. Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature 626, 874–880 (2024).
Leto, D. E. et al. Genome-wide CRISPR analysis identifies substrate-specific conjugation modules in ER-associated degradation. Mol. Cell 73, 377–389.e11 (2019).
Cassidy, K. B., Bang, S., Kurokawa, M. & Gerber, S. A. Direct regulation of Chk1 protein stability by E3 ubiquitin ligase HUWE1. FEBS J. 287, 1985–1999 (2020).
Kunz, V. et al. Targeting of the E3 ubiquitin-protein ligase HUWE1 impairs DNA repair capacity and tumor growth in preclinical multiple myeloma models. Sci. Rep. 10, 18419 (2020).
Thompson, J. W. et al. Quantitative Lys-ϵ-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 289, 28942–28955 (2014).
D’Arca, D. et al. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. USA 107, 5875–5880 (2010).
Poulsen, E. G. et al. HUWE1 and TRIP12 collaborate in degradation of ubiquitin-fusion proteins and misframed ubiquitin. PLoS ONE 7, e50548 (2012).
Hegazi, S. et al. UBR4/POE facilitates secretory trafficking to maintain circadian clock synchrony. Nat. Commun. 13, 1594 (2022).
Hunt, L. C. et al. A key role for the ubiquitin ligase UBR4 in myofiber hypertrophy in drosophila and mice. Cell Rep. 28, 1268–1281.e6 (2019).
Jeong, D. E. et al. Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates. Commun. Biol. 6, 1214 (2023).
Rinschen, M. M. et al. The ubiquitin ligase Ubr4 controls stability of podocin/MEC-2 supercomplexes. Hum. Mol. Genet. 25, 1328–1344 (2016).
Hunkeler, M. et al. Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition. Mol. Cell 81, 3468–3480.e7 (2021).
Grabarczyk, D. B. et al. HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain. Nat. Chem. Biol. 17, 1084–1092 (2021).
Ohtake, F., Tsuchiya, H., Saeki, Y. & Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 115, E1401–E1408 (2018).
Manasanch, E. E. & Orlowski, R. Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433 (2017).
Venkatesan, S. et al. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 29, 563–572 (2018).
Coxon, M. et al. An impaired ubiquitin-proteasome system increases APOBEC3A abundance. NAR Cancer 5, zcad058 (2023).
Shlyakhtenko, L. S., Lushnikov, A. J., Li, M., Harris, R. S. & Lyubchenko, Y. L. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PLoS ONE 9, e99354 (2014).
Chen, X. S. Insights into the structures and multimeric status of apobec proteins involved in viral restriction and other cellular functions. Viruses 13, 497 (2021).
Bohn, M.-F. et al. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure 23, 903–911 (2015).
Aydin, H., Taylor, M. W. & Lee, J. E. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure 22, 668–684 (2014).
Michlits, G. et al. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat. Methods 17, 708–716 (2020).
Schwartz, I. et al. SPOP targets the immune transcription factor IRF1 for proteasomal degradation. Elife 12, e89951 (2023).
Hatziioannou, T., Cowan, S. & Bieniasz, P. D. Capsid-dependent and -independent postentry restriction of primate lentivirus tropism in rodent cells. J. Virol. 78, 1006–1011 (2004).
Yee, J. K., Friedmann, T. & Burns, J. C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43, 99–112 (1994). Pt A.
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Suzuki, K., Bose, P., Leong-Quong, R. Y., Fujita, D. J. & Riabowol, K. REAP: a two minute cell fractionation method. BMC Res. Notes 3, 294 (2010).
Artan, M., Hartl, M., Chen, W. & de Bono, M. Depletion of endogenously biotinylated carboxylases enhances the sensitivity of TurboID-mediated proximity labeling in Caenorhabditis elegans. J. Biol. Chem. 298, 102343 (2022).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
UniProt https://www.uniprot.org/.
R: The R Project for Statistical Computing https://www.r-project.org/.
Madern, M. Cassiopeia_LFQ (2023).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Xie, Z. et al. Gene set knowledge discovery with EnrichR. Curr. Protoc. 1, e90 (2021).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Neuhold, J. et al. GoldenBac: a simple, highly efficient, and widely applicable system for construction of multi-gene expression vectors for use with the baculovirus expression vector system. BMC Biotechnol. 20, 26 (2020).
Ehrmann, J. F. et al. Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex. Science 379, 1117–1123 (2023).
Benjamin, D. et al. Calling somatic SNVs and indels with mutect2. Preprint at https://doi.org/10.1101/861054 (2019).
MutationalPatterns (UMCU Genetics) (2024).
Rozen, S. G. mSigAct. (2024).
Cao, T., Li, Q., Huang, Y. & Li, A. plotnineSeqSuite: a Python package for visualizing sequence data using ggplot2 style. BMC Genomics 24, 585 (2023).
DCC Data Releases | ICGC Data Portal https://dcc.icgc.org/releases/PCAWG.
The Cancer Genome Atlas Program (TCGA) - NCI. https://www.cancer.gov/ccg/research/genome-sequencing/tcga (2022).
nerdgum. menchelab/apobex: Analysis Tools and Code for “Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity” (v1.0). Zenodo. https://doi.org/10.5281/zenodo.17953821 (2025).
Acknowledgements
Next Generation Sequencing analysis was performed by the Vienna Biocenter Core Facilities using the VBCF instrument pool. Proteomics analyses were performed by the Mass Spectrometry Facility at Max Perutz Labs using the VBCF instrument pool; we particularly thank Markus Hartl and WeiQiang Chen for their expert support. Flow cytometry analyses were performed at the BioOptics FACS Facility at the Max Perutz Labs using the Max Perutz Labs instrument pool; we particularly acknowledge Kitti Csalyi, Thomas Sauer, and Johanna Stranner for expert support. Microscopy was performed at the BioOptics Light Microscopy Facility at the Max Perutz Labs; we thank Thomas Peterbauer and Irmgard Fischer for their expert support and training. We thank Johannes Bock for establishing TurboID-related reagents and methodology, Robert Kurzbauer for purification of recombinant proteins, Anna Hakobyan for advice on cancer genome data analysis, Joanna Loizou for expert advice on DNA damage assays, Magdalini Nigritinou and Pablo Araguas-Rodriguez for experimental support, and Marcel Ooms for APOBEC expertise, discussions, and manuscript feedback. We are grateful to the ‘Signaling Mechanisms in Cellular Homeostasis’ doctoral program community, in particular Thomas Decker, Pavel Kovarik and their labs for their technical expertise and help. We thank Life Science Editors for editing services. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This research was funded in whole, or in part, by the Austrian Science Fund (FWF) (grants 10.55776/P36572, 10.55776/P30415, 10.55776/P30231, 10.55776/P36945, 10.55776/F79, and 10.55776/W1261 to G.A.V.). For the purpose of open access, the author has applied a CC-BY public copyright license to any author accepted manuscript version arising from this submission. This work was funded by Austrian Science Fund Special Research Grant (FWF, SFB F79) and an ERC European Union’s Horizon 2020 research and innovation program grant (AdG 694978) to TC, and an Austrian Science Fund Special Research Grant (SFB grant F79) to GEK. VB and S.Sci are the recipients of a DOC fellowship of the Austrian Academy of Sciences. Research at the IMP is supported by Boehringer Ingelheim and the Austrian Research Promotion Agency (Headquarter grant FFG-852936). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization, G.A.V., I.S., V.B., D.H., T.C., and E.K.; Methodology, G.A.V., I.S., V.B., M.M., H.H., Z.H., D.B.G., and J.F.E.; Software, M.M, I.S., and S.S.; Validation, I.S. and V.B.; Formal analysis, I.S., V.B., and M.M.; Investigation, I.S., V.B., and K.H.; Resources, Z.H., D.B.G., J.F.E., and S.Sci.; Data Curation, I.S., V.B., M.M and G.A.V.; Writing—original draft, I.S., V.B. and G.A.V.; Writing—review and editing, I.S., V.B., M.M., H.H., K.H., S.S., Z.H., D.B.G., J.F.E., S.Sci, D.H., J.M., T.C., E.K., and G.A.V.; Visualization, I.S., V.B., and M.M.; Supervision G.A.V.; Project administration, G.A.V.; Funding acquisition, G.A.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schwartz, I., Budroni, V., Meyenberg, M. et al. Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68420-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68420-5