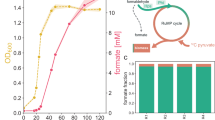
Abstract
Methanol is a promising one-carbon (C1) feedstock for microbial bioconversion; however, engineered Saccharomyces cerevisiae often faces energetic constrains during its assimilation. Here, we develop SC-AOX25, an energy-efficient methylotrophic S. cerevisiae, through engineering of heterologous methanol-formaldehyde-formate (MFF) oxidation pathways coupled with adaptive laboratory evolution. SC-AOX25 efficiently generates adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide (NADH) during methanol metabolism while co-assimilating methanol-derived intermediates (formaldehyde, formate, and CO₂) via native glyoxylate-serine cycle, pentose phosphate pathway, and reductive glycine pathway. Key energy modules - Fdh1sc, Adh2m, Aoxm, and Rgi2m - are characterized for their roles in ATP/NADH synthesis and methylotrophic growth. Formaldehyde-induced DNA-protein crosslinks (DPCs) and large repeated DNA fragments suggest strategies for methanol detoxification and phenotype enhancement. Utilizing SC-AOX25, we enable CO₂ assimilation through non-native Calvin cycle during methanol fermentation, establishing the engineered strain as a robust and energy-efficient methylotrophic platform for further C1 engineering.
Similar content being viewed by others
Data availability
The RNA-seq data and genome resequencing data generated in this study have been deposited in the NCBI SRA database under Bioproject PRJNA1238490 and PRJNA1238044, respectively. The proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE repository under accession PXD064258 and PXD064984. Source data are provided with this paper.
References
Boehm, S. et al. State of Climate Action 2023. World Resour. Inst https://www.wri.org/research/state-climate-action-2023 (2023).
Sollai, S., Porcu, A., Tola, V., Ferrara, F. & Pettinau, A. Renewable methanol production from green hydrogen and captured CO2: a techno-economic assessment. J. CO2 Util. 68, 102345 (2023).
Meunier, N., Chauvy, R., Mouhoubi, S., Thomas, D. & De Weireld, G. Alternative production of methanol from industrial CO2. Renew. Energ. 146, 1192–1203 (2020).
Kelso, P. A., Chow, L. K. M., Carpenter, A. C., Paulsen, I. T. & Williams, T. C. Toward methanol-based biomanufacturing: emerging strategies for engineering synthetic methylotrophy in Saccharomyces cerevisiae. ACS Synth. Biol. 11, 2548–2563 (2022).
Bertau, M., Offermanns, H., Plass, L., Schmidt, F. & Wernicke, H.-J. Methanol: The Basic Chemical and Energy Feedstock of the Future Vol. 1 (Springer, 2014).
Patel, S. K. S. et al. Hierarchical macroporous particles for efficient whole-cell immobilization: Application in bioconversion of greenhouse gases to methanol. ACS Appl. Mater. Interfaces 11, 18968–18977 (2019).
Bennett, R. K., Steinberg, L. M., Chen, W. & Papoutsakis, E. T. Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr. Opin. Biotechnol. 50, 81–93 (2018).
Ochsner, A. M., Sonntag, F., Buchhaupt, M., Schrader, J. & Vorholt, J. A. Methylobacterium extorquens: methylotrophy and biotechnological applications. Appl. Microbiol. Biotechnol. 99, 517–534 (2015).
Mo, X.-H. et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Appl. Microbiol. Biotechnol. 104, 4515–4532 (2020).
Schultenkämper, K., Brito, L. F., López, M. G., Brautaset, T. & Wendisch, V. F. Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus. Appl. Microbiol. Biotechnol. 103, 5879–5889 (2019).
Yang, X., Zheng, Z. & Wang, Y. Bacillus methanolicus: an emerging chassis for low-carbon biomanufacturing. Trends Biotechnol. 43, 274–277 (2025).
Gao, J., Gao, N., Zhai, X. & Zhou, Y. J. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha. iScience 24, 102168 (2021).
Wang, L. et al. Efficient CRISPR–Cas9 mediated multiplex genome editing in yeasts. Biotechnol. Biofuels 11, 277 (2018).
Liu, Q. et al. CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Fact. 18, 144 (2019).
Guo, F., Qiao, Y., Xin, F., Zhang, W. & Jiang, M. Bioconversion of C1 feedstocks for chemical production using Pichia pastoris. Trends Biotechnol. 41, 1066–1079 (2023).
Cai, P., Gao, J. & Zhou, Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Fact. 18, 63 (2019).
Chen, F. Y. H., Jung, H.-W., Tsuei, C.-Y. & Liao, J. C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 182, 933–946.e914 (2020).
Nieh, L.-Y. et al. Evolutionary engineering of methylotrophic E. coli enables fast growth on methanol. Nat. Commun. 15, 8840 (2024).
Reiter, M. A. et al. A synthetic methylotrophic Escherichia coli as a chassis for bioproduction from methanol. Nat. Catal. 7, 560–573 (2024).
Zhan, C. et al. Reprogramming methanol utilization pathways to convert Saccharomyces cerevisiae to a synthetic methylotroph. Nat. Catal. 6, 435–450 (2023).
Guo, Y. et al. Engineering yeasts to co-utilize methanol or formate coupled with CO2 fixation. Metab. Eng. 84, 1–12 (2024).
Espinosa, M. I. et al. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat. Commun. 11, 5564 (2020).
Guo, F. et al. Evolutionary engineering of Saccharomyces cerevisiae: Crafting a synthetic methylotroph via self-reprogramming. Sci. Adv. 10, eadq3484 (2024).
Attfield, P. V. Stress tolerance: The key to effective strains of industrial baker’s yeast. Nat. Biotechnol. 15, 1351–1357 (1997).
Nielsen, J. Yeast systems biology: model organism and cell factory. Biotechnol. J. 14, 1800421 (2019).
Lourens-Hattingh, A. & Viljoen, B. C. Growth and survival of a probiotic yeast in dairy products. Food Res. Int. 34, 791–796 (2001).
Espinosa, M. I., Williams, T. C., Pretorius, I. S. & Paulsen, I. T. Benchmarking two Saccharomyces cerevisiae laboratory strains for growth and transcriptional response to methanol. Syn. Syst. Biotechnol. 4, 180–188 (2019).
Dai, Z. et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour. Technol. 245, 1407–1412 (2017).
Lunt, S. Y. & Vander Heiden, M. G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Bi. 27, 441–464 (2011).
Lieven, C., Herrgård, M. J. & Sonnenschein, N. Microbial methylotrophic metabolism: Recent metabolic modeling efforts and their applications in industrial biotechnology. Biotechnol. J. 13, 1800011 (2018).
Liu, Z., Wang, K., Chen, Y., Tan, T. & Nielsen, J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 3, 274–288 (2020).
Gregory, G. J., Bennett, R. K. & Papoutsakis, E. T. Recent advances toward the bioconversion of methane and methanol in synthetic methylotrophs. Metab. Eng. 71, 99–116 (2022).
Kim, S. et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16, 538–545 (2020).
Yu, H. & Liao, J. C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat. Commun. 9, 3992 (2018).
He, H., Höper, R., Dodenhöft, M., Marlière, P. & Bar-Even, A. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli. Metab. Eng. 60, 1–13 (2020).
Wenk, S. et al. Evolution-assisted engineering of E. coli enables growth on formic acid at ambient CO2 via the serine threonine cycle. Metab. Eng. 88, 14–24 (2025).
Klein, V. J., Irla, M., Gil López, M., Brautaset, T. & Fernandes Brito, L. Unravelling formaldehyde metabolism in bacteria: Road towards synthetic methylotrophy. Microorganisms 10, 220 (2022).
Moioli, E., Mutschler, R. & Züttel, A. Renewable energy storage via CO2 and H2 conversion to methane and methanol: Assessment for small scale applications. Renew. Sust. Enegr. Rev. 107, 497–506 (2019).
Zhu, Z., Kin Tam, T., Sun, F., You, C. & Percival Zhang, Y. H. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway. Nat. Commun. 5, 3026 (2014).
Ullah, A., Hashim, N. A., Rabuni, M. F. & Mohd Junaidi, M. U. A review on methanol as a clean energy carrier: Roles of zeolite in improving production efficiency. Energies 16, 1482 (2023).
Ozimek, P., Kötter, P., Veenhuis, M. & van der Klei, I. J. Hansenula polymorpha and Saccharomyces cerevisiae Pex5p’s recognize different, independent peroxisomal targeting signals in alcohol oxidase. FEBS Lett. 580, 46–50 (2006).
Zhong, W., Li, H. & Wang, Y. Design and construction of artificial biological systems for one-carbon utilization. BioDes. Res. 5, 0021 (2023).
Sundström, M., Lindqvist, Y., Schneider, G., Hellman, U. & Ronne, H. Yeast TKL1 gene encodes a transketolase that is required for efficient glycolysis and biosynthesis of aromatic amino acids. J. Biol. Chem. 268, 24346–24352 (1993).
Navarro-Aviño, J. P., Prasad, R., Miralles, V. J., Benito, R. M. & Serrano, R. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15, 829–842 (1999).
Ruiz-Amil, M., de Torrontegui, G., Palacián, E., Catalina, L. & Losada, M. Properties and function of yeast pyruvate carboxylase. J. Biol. Chem. 240, 3485–3492 (1965).
Qin, N. et al. Increased CO2 fixation enables high carbon-yield production of 3-hydroxypropionic acid in yeast. Nat. Commun. 15, 1591 (2024).
Chandel, N. S. Glycolysis. Cold Spring Harb. Perspect. Biol. 13, a040535 (2021).
Kresnowati, M. T. A. P. et al. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol. Syst. Biol. 2, 49 (2006).
Byrne, K. P. & Wolfe, K. H. The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15, 1456–1461 (2005).
Xia, P. F. et al. Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 6, 276–283 (2017).
Papapetridis, I. et al. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels 11, 17–17 (2018).
Yasokawa, D. et al. Toxicity of methanol and formaldehyde towards Saccharomyces cerevisiae as assessed by DNA microarray analysis. Appl. Biochem. Biotech. 160, 1685–1698 (2010).
Stingele, J. & Jentsch, S. DNA–protein crosslink repair. Nat. Rev. Mol. Cell Bio. 16, 455–460 (2015).
Falcone, C. & Mazzoni, C. External and internal triggers of cell death in yeast. Cell. Mol. Life Sci. 73, 2237–2250 (2016).
Carmona-Gutierrez, D. et al. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 17, 763–773 (2010).
Spégel, C. F. et al. Amperometric response from the glycolytic versus the pentose phosphate pathway in Saccharomyces cerevisiae cells. Anal. Chem. 79, 8919–8926 (2007).
Lu, X. et al. Constructing a synthetic pathway for acetyl-coenzyme a from one-carbon through enzyme design. Nat. Commun. 10, 1378 (2019).
Mao, Y. F. et al. Non-natural aldol reactions enable the design and construction of novel one-carbon assimilation pathways in vitro. Front. Microbiol. 12, 677596 (2021).
Nattermann, M. et al. Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo. Nat. Commun. 14, 2682 (2023).
Henras, A. K., Plisson-Chastang, C., O’Donohue, M.-F., Chakraborty, A. & Gleizes, P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. WIREs RNA 6, 225–242 (2015).
Nomura, M. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 66, 555–565 (2001).
Berger, K. H. & Yaffe, M. P. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol 8, 508–513 (2000).
de Zamaroczy, M. & Bernardi, G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae - a review. Gene 47, 155–177 (1986).
Zhang, Y. et al. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat. Commun. 10, 1053 (2019).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Evers, M. E., Harder, W. & Veenhuis, M. In vitro dissociation and re-assembly of peroxisomal alcohol oxidases of Hansenula polymorpha and Pichia pastoris. FEBS Lett. 368, 293–296 (1995).
Couderc, R. & Baratti, J. Oxidation of methanol by the yeast, Pichia pastoris. purification and properties of the alcohol oxidase. Agric. Biol. Chem. 44, 2279–2289 (1980).
SchÜTte, H., Flossdorf, J., Sahm, H. & Kula, M.-R. Purification and properties of formaldehyde dehydrogenase and formate dehydrogenase from Candida boidinii. Eur. J. Biochem. 62, 151–160 (1976).
Degrassi, G., Uotila, L., Klima, R. & Venturi, V. Purification and properties of an esterase from the yeast Saccharomyces cerevisiae and identification of the encoding gene. Appl. Environ. Microbiol. 65, 3470–3472 (1999).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Overkamp, K. M. et al. Functional analysis of structural genes for NAD+-dependent formate dehydrogenase in Saccharomyces cerevisiae. Yeast 19, 509–520 (2002).
Seah, T. C. M. & Kaplan, J. G. Purification and properties of the catalase of bakers’ yeast. J. Biol. Chem. 248, 2889–2893 (1973).
Ruuska, S. A., Badger, M. R., Andrews, T. J. & von Caemmerer, S. Photosynthetic electron sinks in transgenic tobacco with reduced amounts of Rubisco: little evidence for significant Mehler reaction. J. Exp. Bot. 51, 357–368 (2000).
Yamori, W. & von Caemmerer, S. Effect of Rubisco activase deficiency on the temperature response of CO2 assimilation rate and Rubisco activation state: Insights from transgenic tobacco with reduced amounts of Rubisco activase. Plant Physiol. 151, 2073–2082 (2009).
Sulpice, R. et al. Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ. 30, 1163–1175 (2007).
MacElroy Robert, D., Mack Henry, M. & Johnson Emmett, J. Properties of phosphoribulokinase from Thiobacillus neapolitanus. J. Bacteriol. 112, 532–538 (1972).
Lee, J.-Y., Cheong, D.-E. & Kim, G.-J. A novel assay system for the measurement of transketolase activity using xylulokinase from Saccharomyces cerevisiae. Biotechnol. Lett. 30, 899–904 (2008).
Wang, X., Mann Craig, J., Bai, Y., Ni, L. & Weiner, H. Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J. Bacteriol. 180, 822–830 (1998).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔCT method. Methods 25, 402–408 (2001).
Burda, P. et al. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: Identification of the ALG9 gene encoding a putative mannosyl transferase. Proc. Natl. Acad. Sci. USA 93, 7160–7165 (1996).
Wang, Y., Parsons, L. R. & Su, X. AccuCor2: isotope natural abundance correction for dual-isotope tracer experiments. Lab. Invest. 101, 1403–1410 (2021).
Cota-Sánchez, J. H., Remarchuk, K. & Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 24, 161–167 (2006).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 80–92 (2012).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Shen, S. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. USA 111, E5593–E5601 (2014).
Castell, C. H. & Smith, B. Measurement of formaldehyde in fish muscle using TCA extraction and the Nash reagent. J. Fish. Res. Board Can. 30, 91–98 (1973).
Kamiloglu, S., Sari, G., Ozdal, T. & Capanoglu, E. Guidelines for cell viability assays. Food Front. 1, 332–349 (2020).
Sun, P., Zhang, H., Sun, Y. & Liu, J. The recent development of fluorescent probes for the detection of NADH and NADPH in living cells and in vivo. Spectrochim. Acta A. 245, 118919 (2021).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 14, 513–520 (2017).
Yu, F. et al. Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant. Mol. Cell. Proteom. 19, 1575–1585 (2020).
Flamholz, A., Noor, E., Bar-Even, A. & Milo, R. eQuilibrator-the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775 (2012).
Beber, M. E. et al. eQuilibrator 3.0: a database solution for thermodynamic constant estimation. Nucleic Acids Res. 50, D603–D609 (2022).
Acknowledgements
This work was supported by the National Science Foundation of China (32401209), National Key R&D Program of China (2022YFA0912800), National Natural Science Foundation of China (22178233), Talents Program of Sichuan Province, Fundamental Research Funds for the Central Universities (SCU2025D014), Double First-Class University Plan of Sichuan University, State Key Laboratory of Polymer Materials Engineering (sklpme 2020-03-01), Suzhou Municipal High-Level Talent Entrepreneurship Fund, Tianfu Emei Program of Sichuan Province (2022-EC02-00073-CG) the Center of Synthetic Biology and Integrated Bioengineering (WU2022A006, WU2022A007, WU2023A009), Research Plan for Westlake University Funded Scientific Research Project (WU2022C032), Zhejiang Key Laboratory of Low-Carbon Intelligent Synthetic Biology (2024ZY01025), and Shenzhen Science and Technology Program (RCYX20231211090115013). We thank Wenwen Zhang and Xiaoyan Xu at Mass Spectrometry & Metabolomics Core Facility of Westlake University for MS analysis.
Author information
Authors and Affiliations
Contributions
W.Z. and Y.W. conceived and designed the study. Y.W. supervised the project. W.Z. and N.L. performed the experiments and data processing, and analyzed the data. B.C. completed the analysis of the structure of Aoxm and Adh2m, respectively. H.S. and X.F. were involved in constructing some of the engineered yeast strains needed for the study. W.Z. and Y.W. wrote and revised the manuscript with input of all authors. J.L., J.G., and B.W. contributed to the review, editing, and final approval of the manuscript. They provided valuable insights and suggestions to improve the quality of the research paper.
Corresponding author
Ethics declarations
Competing interests
Authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Wenming Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Declaration of generative AI and AI-ASSISTED technologies
During the preparation of this manuscript, the authors utilized Grammarly (https://www.grammarly.com/) and DeepSeek (www.deepseek.com) for grammatical revision and language polishing. All AI-generated suggestions were critically reviewed and manually adjusted by the authors, who assume full responsibility for the intellectual content and academic integrity of this work.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhong, W., Liu, N., Chen, B. et al. Engineering energy-efficient Saccharomyces cerevisiae for methanol and CO2 assimilation. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68516-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68516-y