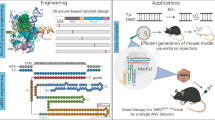
Abstract
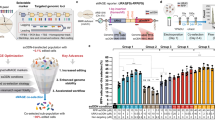
Eukaryotic Fanzor proteins are compact, programmable RNA-guided nucleases with substantial potential for genome editing, although their efficiency in mammalian cells remains suboptimal. Here, we present a combinatorial engineering strategy to optimize a representative Fanzor system, MmeFz2–ωRNA. AlphaFold3-powered rational redesign produced a minimized ωRNA scaffold that is 30% smaller while maintaining up to 82.2% efficiency. Synergistic structure-guided and AI-augmented protein engineering generated two variants, enMmeFz2 and evoMmeFz2, which exhibited an average ~32-fold increase in activity across 38 genomic loci. Moreover, fusion of the non-specific DNA-binding domain HMG-D further enhanced editing performance (enMmeFz2-HMG-D and evoMmeFz2-HMG-D). Notably, evoMmeFz2-HMG-D demonstrated robust in vivo genome editing activity, enabling dystrophin restoration in humanized male Duchenne muscular dystrophy mouse models via single adeno-associated virus (AAV) delivery. This study establishes Fanzor2 as a gene editing platform for genome engineering and therapeutic applications, and underscores the power of AI-guided engineering to accelerate genome editor development while reducing experimental burden.
Similar content being viewed by others
Data availability
Next-generation sequencing data are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive database under the BioProject accession code PRJNA1259048. Source data are provided with this paper.
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Fellmann, C., Gowen, B. G., Lin, P. C., Doudna, J. A. & Corn, J. E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 16, 89–100 (2017).
Chen, K., Wang, Y., Zhang, R., Zhang, H. & Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019).
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018).
Karvelis, T. et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 48, 5016–5023 (2020).
Takeda, S. N. et al. Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell 81, 558–570.e553 (2021).
Wu, Z. et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 17, 1132–1138 (2021).
Wang, Y. et al. Guide RNA engineering enables efficient CRISPR editing with a miniature Syntrophomonas palmitatica Cas12f1 nuclease. Cell Rep. 40, 111418 (2022).
Kong, X. et al. Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing. Nat. Commun. 14, 2046 (2023).
Pausch, P. et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337 (2020).
Pausch, P. et al. DNA interference states of the hypercompact CRISPR-CasΦ effector. Nat. Struct. Mol. Biol. 28, 652–661 (2021).
Carabias, A. et al. Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3. Nat. Commun. 12, 4476 (2021).
Chen, W. et al. Cas12n nucleases, early evolutionary intermediates of type V CRISPR, comprise a distinct family of miniature genome editors. Mol. Cell 83, 2768–2780.e2766 (2023).
Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021).
Nakagawa, R. et al. Cryo-EM structure of the transposon-associated TnpB enzyme. Nature 616, 390–397 (2023).
Sasnauskas, G. et al. TnpB structure reveals minimal functional core of Cas12 nuclease family. Nature 616, 384–389 (2023).
Li, Z. et al. Engineering a transposon-associated TnpB-ωRNA system for efficient gene editing and phenotypic correction of a tyrosinaemia mouse model. Nat. Commun. 15, 831 (2024).
Xiang, G. et al. Evolutionary mining and functional characterization of TnpB nucleases identify efficient miniature genome editors. Nat. Biotechnol. 42, 745–757 (2024).
Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021).
Schuler, G., Hu, C. & Ke, A. Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Science 376, 1476–1481 (2022).
Han, D. et al. Development of miniature base editors using engineered IscB nickase. Nat. Methods 20, 1029–1036 (2023).
Yan, H. et al. Assessing and engineering the IscB-ωRNA system for programmed genome editing. Nat. Chem. Biol. 20, 1617–1628 (2024).
Han, L. et al. Engineering miniature IscB nickase for robust base editing with broad targeting range. Nat. Chem. Biol. 20, 1629–1639 (2024).
Xue, N. et al. Engineering IscB to develop highly efficient miniature editing tools in mammalian cells and embryos. Mol. Cell 84, 3128–3140.e3124 (2024).
Xiao, Q. et al. Engineered IscB-ωRNA system with expanded target range for base editing. Nat. Chem. Biol. 21, 100–108 (2025).
Saito, M. et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 620, 660–668 (2023).
Jiang, K. et al. Programmable RNA-guided DNA endonucleases are widespread in eukaryotes and their viruses. Sci. Adv. 9, eadk0171 (2023).
Xu, P. et al. Structural insights into the diversity and DNA cleavage mechanism of Fanzor. Cell 187, 5238–5252.e20 (2024).
Jiang, K. et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science 387, eadr6006 (2025).
Hennig, J. Structural biology of RNA and protein-RNA complexes after alphaFold3. Chembiochem 26, e202401047 (2025).
Su, M. et al. Molecular basis and engineering of miniature Cas12f with C-rich PAM specificity. Nat. Chem. Biol. 20, 180–189 (2024).
Kim, D. Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102 (2022).
Gao, Z., Herrera-Carrillo, E. & Berkhout, B. Delineation of the exact transcription termination signal for type 3 polymerase III. Mol. Ther. Nucleic Acids 10, 36–44 (2018).
Zhang, X. et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 22, 740–750 (2020).
Yin, S. et al. Engineering of efficiency-enhanced Cas9 and base editors with improved gene therapy efficacies. Mol. Ther. 31, 744–759 (2023).
Yang, C. et al. HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing. Nat. Commun. 14, 2430 (2023).
Ding, X. et al. Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating Peptides. CRISPR J. 2, 51–63 (2019).
Yin, J. et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat. Commun. 13, 1204 (2022).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Roberts, T. C., Wood, M. J. A. & Davies, K. E. Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug Discov. 22, 917–934 (2023).
Flanigan, K. M. et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum. Mutat. 30, 1657–1666 (2009).
Min, Y. L., Bassel-Duby, R. & Olson, E. N. CRISPR correction of Duchenne muscular dystrophy. Annu. Rev. Med. 70, 239–255 (2019).
Lin, J. et al. Adenine base editing-mediated exon skipping restores dystrophin in humanized Duchenne mouse model. Nat. Commun. 15, 5927 (2024).
Bao, W. & Jurka, J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob. DNA 4, 12 (2013).
Altae-Tran, H. et al. Diversity, evolution, and classification of the RNA-guided nucleases TnpB and Cas12. Proc. Natl. Acad. Sci. USA 120, e2308224120 (2023).
Dong, C., Gou, Y. & Lian, J. SgRNA engineering for improved genome editing and expanded functional assays. Curr. Opin. Biotechnol. 75, 102697 (2022).
Wu, Z. et al. Structure and engineering of miniature Acidibacillus sulfuroxidans Cas12f1. Nat. Catal. 6, 695–709 (2023).
Acknowledgements
We are grateful for the support from the Gene Editing Scientific Teaching (NWAFU-GEST), High-Performance Computing (HPC), and Life Science Research Core Service platforms (K.R. Huang, X.R. Liu, L. Chen, M. Zhou, and L.Q. Li) at Northwest A&F University (NWAFU). The authors also wish to express their gratitude to the members of HuidaGene Therapeutics Co., Ltd. for their contributions in supplying experimental materials and insightful discussions. This work is supported by the National Natural Science Foundation of China (32441080, 32301251 to Y.W. and 22207074 to Z.W.), the Biological Breeding-Major Projects (2023ZD04074 to K.X., 2023ZD04051 to Y.W., and 2022ZD04014 to X.W.), the National Key Research and Development Program of China (2023YFF1000904 to X.W.), the National Science and Technology Major Project of China (2023ZD0500500 to Z.W.), the China Agricultural Research System (CARS-39-03 to X.W.), and local grants (2024A02004-1-3, 2025NC-YBXM-109, and QCYRCXM-2023-104 to Y.W. and 2023A02011-2 to X.W.).
Author information
Authors and Affiliations
Contributions
Y.W., Z.W.W., K.X., and X.W. conceived the project. Y.W., Z.W.W., K.X., and X.L. designed the experiments. Y.W., S.L., P.G., and G.L. performed data analysis. Z.W.W. conducted the structural prediction analysis. S. L., P.G., Z.M.W., Y.Y., H.J., Y.F.C., Z.L., B.Z., and M.Z. performed cell transfection and FACS. Y.W. and G.L. performed animal experiments. Y.W. and Z.W.W. wrote the manuscripts. Y.W., X.W., Z.W.W., Y.L.C., K.X., and X.L. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chunyi Hu, Jun-Jie (Gogo) Liu and Hidetoshi Sakurai for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, S., Xu, K., Li, G. et al. Engineering the MmeFz2-ωRNA system for efficient genome editing through an integrated computational-experimental framework. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68644-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68644-5