Abstract
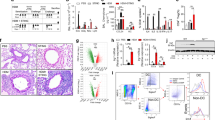
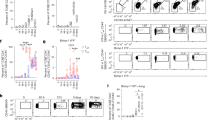
The mechanisms of airway allergen sensing and type 2 immune response initiation remain poorly understood. Using a mouse house dust mite (HDM)-induced allergic airway model, we identify a population of lung macrophages located close to alveolar capillaries that express Ly6G and the nuclear receptor Nr4a1/Nur77. These atypical Ly6G+Nur77+ macrophages preferentially capture airway-delivered allergens and play an important role in initiating HDM-driven T helper type 2 (Th2) responses. They sense the major HDM allergen, the cysteine protease Der p 1, via protease-activated receptor 2 (PAR2), and their activation and accumulation require both PAR2 and Nr4a1/Nur77. These Ly6G+Nur77+ macrophages regulate the migration of conventional migratory dendritic cells (mDCs) to draining mediastinal lymph nodes (mLNs) through cysteinyl leukotriene (CysLT) production, which enhances mDC migration toward CCL21 for T cell priming. Inhibiting CysLT biosynthesis reduces mDC migration and dampens Th2 allergic responses, highlighting possible therapeutic avenues in type 2 immunity.
Similar content being viewed by others
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession GSE291081, and scRNA-seq data have been deposited in GEO under accessions GSE289548 and GSE305119 and are publicly available. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon request. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Code availability
The code used to analyze the scRNA-seq data can be accessed at https://doi.org/10.5281/zenodo.15149503 and https://doi.org/10.5281/zenodo.16848446.
References
Li, D. & Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target Ther. 6, 291 (2021).
Meloun, A. & Leon, B. Sensing of protease activity as a triggering mechanism of Th2 cell immunity and allergic disease. Front. Allergy 4, 1265049 (2023).
Trompette, A. et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature 457, 585–588 (2009).
Hammad, H. et al. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 15, 410–416 (2009).
Bachus, H. et al. Impaired tumor-necrosis-factor-alpha-driven dendritic cell activation limits lipopolysaccharide-induced protection from allergic inflammation in infants. Immunity 50, 225–240 e224 (2019).
Kaur, K. et al. GM-CSF production by non-classical monocytes controls antagonistic LPS-driven functions in allergic inflammation. Cell Rep. 37, 110178 (2021).
Bachus, H. et al. IL-6 prevents Th2 cell polarization by promoting SOCS3-dependent suppression of IL-2 signaling. Cell Mol. Immunol. 20, 651–665 (2023).
Leon, B. Understanding the development of Th2 cell-driven allergic airway disease in early life. Front. Allergy 3, 1080153 (2022).
Leon, B. A model of Th2 differentiation based on polarizing cytokine repression. Trends Immunol. 44, 399–407 (2023).
Serhan, N. et al. House dust mites activate nociceptor-mast cell clusters to drive type 2 skin inflammation. Nat. Immunol. 20, 1435–1443 (2019).
Gurram, R. K. et al. Crosstalk between ILC2s and Th2 cells varies among mouse models. Cell Rep. 42, 112073 (2023).
Halim, T. Y. et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 40, 425–435 (2014).
Tang, H. et al. The T helper type 2 response to cysteine proteases requires dendritic cell-basophil cooperation via ROS-mediated signaling. Nat. Immunol. 11, 608–617 (2010).
Chevigne, A. & Jacquet, A. Emerging roles of the protease allergen Der p 1 in house dust mite-induced airway inflammation. J. Allergy Clin. Immunol. 142, 398–400 (2018).
Chua, K. Y. et al. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. J. Exp. Med 167, 175–182 (1988).
Ballesteros-Tato, A. et al. T follicular helper cell plasticity shapes pathogenic T helper 2 cell-mediated immunity to inhaled house dust mite. Immunity 44, 259–273 (2016).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525 e1519 (2019).
Mildner, A. et al. Genomic characterization of murine monocytes reveals C/EBPbeta transcription factor dependence of Ly6C(-) cells. Immunity 46, 849–862 e847 (2017).
Wang, Z. et al. An immune cell atlas reveals the dynamics of human macrophage specification during prenatal development. Cell 186, 4454–4471 e4419 (2023).
Thomas, G. D. et al. Deleting an Nr4a1 super-enhancer subdomain ablates Ly6C(low) monocytes while preserving macrophage gene function. Immunity 45, 975–987 (2016).
Meloun, A. & León, B. Beyond CCR7: dendritic cell migration in type 2 inflammation. Frontiers in Immunology https://doi.org/10.3389/fimmu.2025.1558228 (2025).
Drazen, J. M., Israel, E. & O’Byrne, P. M. Treatment of asthma with drugs modifying the leukotriene pathway. N. Engl. J. Med 340, 197–206 (1999).
Gusach, A. et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat. Commun. 10, 5573 (2019).
Radauer, C., Bublin, M., Wagner, S., Mari, A. & Breiteneder, H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 121, 847–852 e847 (2008).
Yike, I. Fungal proteases and their pathophysiological effects. Mycopathologia 171, 299–323 (2011).
Wiesner, D. L. et al. Club cell TRPV4 serves as a damage sensor driving lung allergic inflammation. Cell Host Microbe 27, 614–628 e616 (2020).
Schmidlin, F. et al. Protease-activated receptor 2 mediates eosinophil infiltration and hyperreactivity in allergic inflammation of the airway. J. Immunol. 169, 5315–5321 (2002).
Page, K., Ledford, J. R., Zhou, P., Dienger, K. & Wills-Karp, M. Mucosal sensitization to German cockroach involves protease-activated receptor-2. Respir. Res 11, 62 (2010).
Shi, T., Denney, L., An, H., Ho, L. P. & Zheng, Y. Alveolar and lung interstitial macrophages: definitions, functions, and roles in lung fibrosis. J. Leukoc. Biol. 110, 107–114 (2021).
Satoh, T. et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017).
Ruscitti, C. et al. Recruited atypical Ly6G(+) macrophages license alveolar regeneration after lung injury. Sci. Immunol. 9, eado1227 (2024).
Worbs, T., Hammerschmidt, S. I. & Forster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 17, 30–48 (2017).
Leon, B. & Lund, F. E. Compartmentalization of dendritic cell and T-cell interactions in the lymph node: anatomy of T-cell fate decisions. Immunol. Rev. 289, 84–100 (2019).
Alraies, Z. et al. Cell shape sensing licenses dendritic cells for homeostatic migration to lymph nodes. Nat. Immunol. 25, 1193–1206 (2024).
Penteado, L. A. et al. Distinctive role of efferocytosis in dendritic cell maturation and migration in sterile or infectious conditions. Immunology 151, 304–313 (2017).
Bosteels, V. et al. LXR signaling controls homeostatic dendritic cell maturation. Sci. Immunol. 8, eadd3955 (2023).
Hauser, M. A. et al. Inflammation-induced CCR7 oligomers form scaffolds to integrate distinct signaling pathways for efficient cell migration. Immunity 44, 59–72 (2016).
Marsland, B. J. et al. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity 22, 493–505 (2005).
Guak, H. et al. Glycolytic metabolism is essential for CCR7 oligomerization and dendritic cell migration. Nat. Commun. 9, 2463 (2018).
Liu, J. et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1alpha-mediated glycolysis. Immunity 50, 600–615 e615 (2019).
Leon, B. et al. Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat. Immunol. 13, 681–690 (2012).
Lyons-Cohen, M. R., Shamskhou, E. A. & Gerner, M. Y. Site-specific regulation of Th2 differentiation within lymph node microenvironments. J. Exp. Med. https://doi.org/10.1084/jem.20231282 (2024).
He, K. et al. Spatial microniches of IL-2 combine with IL-10 to drive lung migratory T(H)2 cells in response to inhaled allergen. Nat. Immunol. 25, 2124–2139 (2024).
Baptista, A. P. et al. The chemoattractant receptor Ebi2 drives intranodal naive CD4(+) T cell peripheralization to promote effective adaptive immunity. Immunity 50, 1188–1201 e1186 (2019).
Doherty, T. A. et al. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J. Allergy Clin. Immunol. 132, 205–213 (2013).
Salimi, M. et al. Cysteinyl leukotriene E(4) activates human group 2 innate lymphoid cells and enhances the effect of prostaglandin D(2) and epithelial cytokines. J. Allergy Clin. Immunol. 140, 1090–1100 e1011 (2017).
McGinty, J. W. et al. Tuft-cell-derived leukotrienes drive rapid anti-helminth immunity in the small intestine but are dispensable for anti-protist immunity. Immunity 52, 528–541 e527 (2020).
Ualiyeva, S. et al. Tuft cell-produced cysteinyl leukotrienes and IL-25 synergistically initiate lung type 2 inflammation. Sci. Immunol. 6, eabj0474 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chen, Y., Chen, L., Lun, A. T. L., Baldoni, P. L. & Smyth, G. K. edgeR v4: powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaf018 (2025).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Andreatta, M. & Carmona, S. J. UCell: Robust and scalable single-cell gene signature scoring. Comput Struct. Biotechnol. J. 19, 3796–3798 (2021).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Gu, Z. Complex heatmap visualization. Imeta 1, e43 (2022).
Acknowledgements
We thank Dr. Catherine C. Hedrick (Augusta University, Augusta, GA, USA), Pamela A. Frischmeyer-Guerrerio, Karen Laky, and Justin Lack (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA) for providing mice and bioinformatics support. We also thank Becca Burnham, Kelsey Browning, and Thomas ‘Scott’ Simpler (University of Alabama at Birmingham, Birmingham, AL, USA) for animal husbandry, and the UAB FCSC Core for assistance with cell sorting and preparation of scRNA-seq libraries. This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov). This research was supported in part by National Institutes of Health (NIH) grant 2R01AI116584 to BL, and by the Intramural Research Program of the NIH. The contributions of the NIH author(s) are considered Works of the United States Government. The findings and conclusions presented in this paper are those of the author(s) and do not necessarily reflect the views of the NIH or the U.S. Department of Health and Human Services.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.M. and B.L.; Methodology, A.M., H.B., C.L., S.D., G.P., J.C.G., and B.L.; Formal analysis, A.M., B.D., D.D.H., P.L., A.F.R., and B.L.; Visualization, A.M., B.D., and B.L.; Writing – original draft, A.M. and B.L.; Writing – review and editing, B.L.; Supervision, B.L.; Funding acquisition, B.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Barbara Balestrieri, Masahiro Kiuchi and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meloun, A., Bachus, H., Lewis, C. et al. Atypical pericapillary Ly6G⁺Nur77⁺ macrophages initiate type-2 immune responses to allergens in the mouse lung. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68652-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68652-5