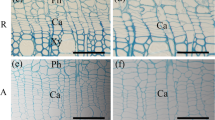
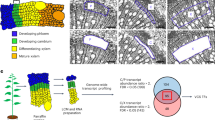
Abstract
Secondary vascular growth is a conserved mechanism that gives rise to vascular tissues produced via a single vascular cambium. Molecular mechanisms underlying this process are characterized mainly in model species with typical vascular architectures, while the genetics underlying ecologically-important atypical vascular architectures remain unexplored. We use developmental anatomy, comparative transcriptomics, molecular evolutionary analyses, and heterologous gene expression to address this knowledge gap, investigating how multiple ectopic cambia (EC) form in the woody vine Japanese wisteria. Anatomical studies show EC in Japanese wisteria arise from cortical parenchyma, and cambium-specific transcriptome comparisons reveal that genes acting as regulators of typical cambium development in model species are likewise associated with EC development. Gene trees of KNOX genes suggest that duplication events may contribute to EC formation, including a Fabaceae-specific duplication of KNAT2/6, which is detected as being under positive selection. We also demonstrate that KNOX genes from Japanese wisteria show canonical KNOX-like activity in heterologous functional assays, although no vascular aberrations were observed. Overall, these findings provide the first insights into the genetics of EC formation in “natural variants”, advancing our understanding of the molecular mechanisms regulating vascular variants in seed plants.
Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are publicly available. The RNA sequencing data are available in SRA [SRR28389658], [SRR29802342-SRR29802358]. Other datasets generated in this study have been deposited in the GitHub database (Cunha-Neto et al.87; https://github.com/anthonysnead/Fabaceae-Ectopic_Cambia_Transcriptomics) and Figshare (Cunha-Neto et al.88; https://doi.org/10.6084/m9.figshare.26524381). Source data are provided with this paper.
Code availability
A custom R script was written for gene expression analyses and is available on GitHub (https://github.com/anthonysnead/Fabaceae-Ectopic_Cambia_Transcriptomics).
References
Hunziker, P. & Greb, T. Stem cells and differentiation in vascular tissues. Annu. Rev. Plant Biol. 75, 11.1–11.27 (2024).
Lanner, R. M. Why do trees live so long? Ageing Res. Rev. 1, 653–671 (2002).
Sillett, S. C. et al. Increasing wood production through old age in tall trees. For. Ecol. Manag. 259, 976–994 (2010).
Rowe, N. P. & Speck, T. The evolution of angiosperm lianescence: a perspective from xylem structure-function. in Ecology of lianas. 221–250 (JohnWiley & Sons, Chichester, 2015).
Fischer, J. B. & Ewers, F. W. Structural responses to stem injury in vines. in The Biology of Vines. 99–124 (Cambridge University Press, Cambridge, 1991).
Cunha-Neto, I. L. Vascular variants in seed plants—a developmental perspective. AoB PLANTS 15, 1–15 (2023).
Cunha-Neto, I. L. & Onyenedum, J. G. Ectopic cambia: connections between natural and experimental vascular mutants. Am. J. Bot. 110, e16246 (2023).
Terrazas, T., Aguilar-Rodríguez, S. & Ojanguren, C. T. Development of successive cambia, cambial activity, and their relationship to physiological traits in Ipomoea arborescens (Convolvulaceae) seedlings. Am. J. Bot. 98, 765–774 (2011).
Robert, E. M. R. et al. Successive cambia: a developmental oddity or an adaptive structure? PLoS ONE 6, e16558 (2011).
Nejapa, R., Cabanillas, P. A. & Pace, M. R. Cortical origin of the successive cambia in the stems of the charismatic temperate lianescent genus Wisteria (Fabaceae) and its systematic importance. Bot. J. Linn. Soc. 199, 667–677 (2022).
Cunha-Neto, I. L. & Onyenedum, J. G. How to climb and grow stronger: lessons from ornamental wisterias. Arnoldia 80, 38–47 (2023).
Trusty, J. L., Lockaby, B. G., Zipperer, W. C. & Goertzen, L. R. Horticulture, hybrid cultivars and exotic plant invasion: a case study of Wisteria (Fabaceae). Bot. J. Linn. Soc. 158, 593–601 (2008).
Tamaio, N., Vieira, R. C. & Angyalossy, V. Origin of successive cambia on the stem in three species of Menispermaceae. Rev. bras. Bot. 32, 839–848 (2009).
Turley, E. K. & Etchells, J. P. Laying it on thick: a study in secondary growth. J. Exp. Bot. 73, 665–679 (2022).
Groover, A. The vascular cambium revisited. IAWA J. 44, 531–538 (2023).
Robischon, M., Du, J., Miura, E. & Groover, A. The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 155, 1214–1225 (2011).
Zhu, Y., Song, D., Sun, J., Wang, X. & Li, L. PtrHB7, a class III HD-zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 6, 1331–1343 (2013).
Zhu, Y., Song, D., Xu, P., Sun, J. & Li, L. An HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 16, 808–817 (2018).
Kucukoglu, M., Nilsson, J., Zheng, B., Chaabouni, S. & Nilsson, O. WUSCHEL - RELATED HOMEOBOX 4 (WOX 4) -like genes regulate cambial cell division activity and secondary growth in Populus trees. N. Phytol. 215, 642–657 (2017).
Zhang, J. et al. Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants 5, 1033–1042 (2019).
Zhang, J. et al. Molecular features of secondary vascular tissue regeneration after bark girdling in Populus. N. Phytol. 192, 869–884 (2011).
Zhang, Y., Wang, X., Zhang, J. & He, X.-Q. Plant in situ tissue regeneration: dynamics, mechanisms and implications for forestry research. For. Res. 3, 3–8 (2023).
Schrader, J. et al. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16, 2278–2292 (2004).
Bryant, D. M. et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776 (2017).
UniProt Consortium UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 51, D523–D531 (2023).
Hoffman, G. E. & Roussos, P. Dream: powerful differential expression analysis for repeated measures designs. Bioinformatics 37, 192–201 (2021).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Hay, A. & Tsiantis, M. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165 (2010).
Ma, J., Mei, G., Liu, H. & Li, H. Overexpression of a novel LcKNOX transcription factor from liriodendron chinense induces lobed leaves in arabidopsis thaliana. Forests 11, 33 (2020).
Larsson, E., Sitbon, F. & von Arnold, S. Differential regulation of Knotted1-like genes during establishment of the shoot apical meristem in Norway spruce (Picea abies). Plant Cell Rep. 31, 1053–1060 (2012).
Shani, E. et al. Stage-specific regulation of solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21, 3078–3092 (2009).
Zhuang, Y. et al. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat. Plants 8, 233–244 (2022).
Landis, J. B. et al. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 105, 348–363 (2018).
Zhang, T. et al. Phylogenomic profiles of whole-genome duplications in Poaceae and landscape of differential duplicate retention and losses among major Poaceae lineages. Nat. Commun. 15, 3305 (2024).
Terrazas, T. Origin and activity of successive cambia in cycas (Cycadales). Am. J. Bot. 78, 1335–1344 (1991).
Carlquist, S. Wood anatomy of Gnetales in a functional, ecological, and evolutionary context. Aliso https://doi.org/10.5642/aliso.20123001.05 (2012).
Groover, A. T. et al. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61, 917–932 (2006).
Du, J., Mansfield, S. D. & Groover, A. T. The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J. 60, 1000–1014 (2009).
Liebsch, D. et al. Class I KNOX transcription factors promote differentiation of cambial derivatives into xylem fibers in the Arabidopsis hypocotyl. Development 141, 4311–4319 (2014).
Byrne, M. E. et al. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971 (2000).
Semiarti, E. et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates the formation of a symmetric lamina, the establishment of venation and the repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783 (2001).
Byrne, M. E., Simorowski, J. & Martienssen, R. A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965 (2002).
Woerlen, N. et al. Repression of BLADE-ON-PETIOLE genes by KNOX homeodomain protein BREVIPEDICELLUS is essential for differentiation of secondary xylem in Arabidopsis root. Planta 245, 1079–1090 (2017).
Kim, M.-H. et al. Wood transcriptome profiling identifies critical pathway genes of secondary wall biosynthesis and novel regulators for vascular cambium development in Populus. Genes 10, 690 (2019).
Zhao, Y. et al. KNAT 2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. N. Phytol. 225, 1531–1544 (2020).
Smith, H. M. S. & Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15, 1717–1727 (2003).
Khan, M. et al. Antagonistic Interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 158, 946–960 (2012).
Ragni, L., Belles-Boix, E., Günl, M. & Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis Inflorescences. Plant Cell 20, 888–900 (2008).
Champagne, C. E. M. et al. Compound leaf development and evolution in the legumes. Plant Cell 19, 3369–3378 (2007).
Johnson, L. A. & Douglas, C. J. Populus trichocarpa MONOPTEROS/AUXIN RESPONSE FACTOR5 (ARF5) genes: comparative structure, sub-functionalization, and Populus – Arabidopsis microsynteny. This article is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Can. J. Bot. 85, 1058–1070 (2007).
Smetana, O. et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489 (2019).
Chen, J. et al. Differential regulation of auxin and cytokinin during the secondary vascular tissue regeneration in Populus trees. N. Phytol. 224, 188–201 (2019).
Etchells, J. P., Provost, C. M. & Turner, S. R. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet 8, e1002997 (2012).
Wybouw, B., Zhang, X. & Mähönen, A. P. Vascular cambium stem cells: past, present and future. N. Phytol. 243, 851–865 (2024).
Dai, X. et al. Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa. Nat. Plants 9, 96–111 (2023).
Groover, A. Roles for epigenetics in wood formation and stress response in trees–from basic biology to forest management. Front. Epigenet. Epigenom. https://doi.org/10.3389/freae.2025.1476499 (2025).
Lima, A. C. et al. Liana's attachment to supports leads to profound changes in xylem anatomy and transcriptional profile of cambium and differentiating xylem. Plant, Cell Environ. 47, 5172–5188 (2024).
Plunkert, M. L., Martínez-Gómez, J., Madrigal, Y., Hernández, A. I. & Tribble, C. M. Tuber, or not tuber: molecular and morphological basis of underground storage organ development. Curr. Opin. Plant Biol. 80, 102544 (2024).
Tribble, C. M., Martínez-Gómez, J., Alzate-Guarín, F., Rothfels, C. J. & Specht, C. D. Comparative transcriptomics of a monocotyledonous geophyte reveals shared molecular mechanisms of underground storage organ formation. Evol. Dev. 23, 155–173 (2021).
Barbosa, A. C. F., Pace, M. R., Witovisk, L. & Angyalossy, V. A new method to obtain good anatomical slides of heterogeneous plant parts. IAWA J. 31, 373–383 (2010).
Qiu, Z. et al. Genome-wide analysis reveals dynamic changes in expression of microRNAs during vascular cambium development in Chinese fir, Cunninghamia lanceolata. J. Exp. Bot. 66, 3041–3054 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Chikhi, R. & Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30, 31–37 (2014).
Rivera-Vicéns, R. E., Garcia-Escudero, C. A., Conci, N., Eitel, M. & Wörheide, G. TransPi-a comprehensive Transcriptome Analysis Pipeline for de novo transcriptome assembly. Mol. Ecol. Resour. 22, 2070–2086 (2022).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Davidson, N. M. & Oshlack, A. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 15, 410 (2014).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Li, H., Jiang, F., Wu, P., Wang, K. & Cao, Y. A high-quality genome sequence of model legume lotus japonicus (MG-20) provides insights into the evolution of root nodule symbiosis. Genes 11, 483 (2020).
Tang, H. et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15, 312 (2014).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Hoffman, G. E. & Schadt, E. E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinforma. 17, 483 (2016).
Phillips, H. R., Landis, J. B. & Specht, C. D. Revisiting floral fusion: the evolution and molecular basis of a developmental innovation. J. Exp. Bot. 71, 3390–3404 (2020).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Grabherr, M. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652 (2011).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinforma. 10, 421 (2009).
Aparicio-Puerta, E. et al. sRNAbench and sRNAtoolbox 2022 update: accurate miRNA and sncRNA profiling for model and non-model organisms. Nucleic Acids Res. 50, W710–W717 (2022).
Jin, Y. & Qian, H. V. PhyloMaker2: an updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Divers. 44, 335–339 (2022).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Ranwez, V., Harispe, S., Delsuc, F. & Douzery, E. J. P. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLOS ONE 6, e22594 (2011).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T.-Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Cunha-Neto, I. L., Snead, A. A., Landis, J. B., Callery, N. I. & Husbands, A. Y. et al. Ectopic cambia in Japanese wisteria (Wisteria floribunda) vines are associated with the expression of conserved KNOX genes. GitHub https://doi.org/10.5281/zenodo.18200460 (2026).
Cunha-Neto, I. L., Snead, A. A., Landis, J. B., Callery, N. I. & Husbands, A. Y. et al. Ectopic cambia in Japanese wisteria (Wisteria floribunda) vines are associated with the expression of conserved KNOX genes. Figshare https://doi.org/10.6084/m9.figshare.26524381 (2026).
Acknowledgments
We thank undergraduate research technician Danielle C. Sonnenleiter (Cornell University) for assistance in data collection, past and current members of the Onyenedum lab for continuous feedback, especially Angelique A. Acevedo, for assistance in growing the bean plants, and Mariane S. S. Baena for insightful discussions. We also thank Jocelyn Rose’s lab at Cornell University for allowing access to the cryostat. We want to acknowledge the Arnold Arboretum of Harvard University for providing access to the living collections and financial support through a Sargent Award for Visiting Scholars (I.L.C.N.). This work was supported in part through the NYU IT High Performance Computing resources, services, and staff expertise, as well as the Boyce Thompson Institute’s Computational Biology Center and Cornell’s BioHPC. This work was funded by startup laboratory funds from Cornell University’s College of Agriculture and Life Sciences and New York University, and NSF 2401675 to J.G.O., and startup laboratory funds from Florida International University to I.L.C.N.
Author information
Authors and Affiliations
Contributions
Conceptualization: I.L.C.-N. J.G.O. Methodology: I.L.C.-N., A.A.S., J.B.L., N.I.C., A.Y.H., C.D.S, and J.G.O. Investigation: I.L.C.-N., J.B.L., A.A.S., N.I.C., A.Y.H., and J.G.O. Data curation: I.L.C.-N., A.A.S., and A.Y.H. Writing – Original Draft: I.L.C.-N., J.B.L., A.A.S., and J.G.O. Writing – Review & Editing: I.L.C.-N., A.A.S., J.B.L., N.I.C., A.Y.H., C.D.S., and J.G.O.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cunha-Neto, I.L., Snead, A.A., Landis, J.B. et al. Ectopic cambia in wisteria vines are associated with the expression of conserved KNOX genes. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68669-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68669-w