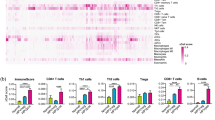
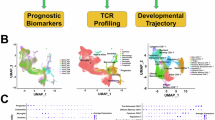
Abstract
Hypopharyngeal squamous cell carcinoma (HPSCC), an aggressive head and neck cancer with dismal prognosis, faces persistent chemoresistance to standard TPF (docetaxel, cisplatin, 5-fluorouracil) regimen. However, the immunological mechanisms underlying chemoresistance remain undefined. Here, we perform longitudinal single-cell RNA sequencing (scRNA-seq) profiling of paired pre-/post-TPF HPSCC specimens, mapping immune cell dynamics underlying chemoresistance. Our study identifies ZNF683+ natural killer (NK) cells as a gatekeeper of chemotherapy efficacy through integrated single-cell transcriptomics, spatial multiplex immunohistochemistry and functional validation. Moreover, pretreatment baseline enrichment of ZNF683+ NK cells predicts TPF response, while GZMK+CD8+ effector memory T cells function as the predominant immunologic effector to successful TPF intervention. Mechanistically, bioinformatics and in vitro coculture data reveal that ZNF683+ NK cells directly interact with CD8+ T cells, and drive an MHC-I-dependent licensing of polyfunctional GZMK+CD8+ effector memory T cells. Collectively, this NK-CD8+ axis provides a potential predictive biomarker and therapeutic target to overcome chemoresistance in patients with HPSCC.
Similar content being viewed by others
Data availability
The sequencing data that support the findings of this study have been deposited in the China National Center for Bioinformation (CNCB-NGDC): https://ngdc.cncb.ac.cn/gsa-human/browse/HRA012395 (Project Number: HRA012395; BioProject ID: PRJCA043247). The data that support the findings of this study are available from the authors on reasonable request. To request access, please contact: Corresponding Author: Yong Liu (liuyongent@csu.edu.cn), First Author: Guo Li (liguoent@csu.edu.cn). Access is granted to qualified researchers for academic and non-commercial research purposes only. Once a data request is approved, an access link will be provided within approximately one week. The standard access period granted is two weeks, which can be adjusted based on the requester’s project needs. Source data are provided with this paper.
References
Garneau, J. C., Bakst, R. L. & Miles, B. A. Hypopharyngeal cancer: a state of the art review. Oral. Oncol. 86, 244–250 (2018).
Chiesa-Estomba, C. M., et al. Radiomics in hypopharyngeal cancer management: a state-of-the-art review. Biomedicines 11, 805 (2023).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Zhang, Y. et al. Integrated transcriptome study of the tumor microenvironment for treatment response prediction in male predominant hypopharyngeal carcinoma. Nat. Commun. 14, 1466 (2023).
Li, R. et al. Induction chemotherapy of modified docetaxel, cisplatin, 5-fluorouracil for laryngeal preservation in locally advanced hypopharyngeal squamous cell carcinoma. Head. Neck 44, 2018–2029 (2022).
Merlano, M. C. et al. Phase III randomized study of induction chemotherapy followed by definitive radiotherapy + cetuximab versus chemoradiotherapy in squamous cell carcinoma of the head and neck: The INTERCEPTOR-GONO Study (NCT00999700). Oncology 98, 763–770 (2020).
Tian, Y. et al. Efficacy and safety of anti-EGFR agents administered concurrently with standard therapies for patients with head and neck squamous cell carcinoma: a systematic review and meta-analysis of randomized controlled trials. Int J. Cancer 142, 2198–2206 (2018).
Harrington, K. J. et al. Efficacy and safety of nivolumab plus ipilimumab vs nivolumab alone for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck: the phase 2 checkmate 714 randomized clinical trial. JAMA Oncol. 9, 779–789 (2023).
Bozec, A. et al. Induction chemotherapy-based larynx preservation program for locally advanced hypopharyngeal cancer: oncologic and functional outcomes and prognostic factors. Eur. Arch. Otorhinolaryngol. 273, 3299–3306 (2016).
Gomes, I. N. F., et al. Comprehensive molecular landscape of cetuximab resistance in head and neck cancer cell lines. Cells 11, 154 (2022).
Leemans, C. R., Snijders, P. J. F. & Brakenhoff, R. H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269–282 (2018).
Wei, J. et al. CD56 dim+ immune infiltration and reprogrammed TMEs associated with response to neoadjuvant anti-PD-1 immunotherapy plus concurrent chemoradiotherapy in locally advanced gastric or gastroesophageal junction adenocarcinoma. Cancer Res. 82, 6176–6176 (2022).
Li, J. et al. Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell 41, 1152–1169 e1157 (2023).
Tang, F. et al. A pan-cancer single-cell panorama of human natural killer cells. Cell 186, 4235–4251 e4220 (2023).
Ma, J. et al. A blueprint for tumor-infiltrating B cells across human cancers. Science 384, eadj4857 (2024).
Marquardt, N. et al. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat. Commun. 10, 3841 (2019).
Liu, Y. et al. Tumour heterogeneity and intercellular networks of nasopharyngeal carcinoma at single cell resolution. Nat. Commun. 12, 741 (2021).
Zheng, Y. et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat. Commun. 11, 6268 (2020).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Liu, Z. et al. Comprehensive analysis of myeloid signature genes in head and neck squamous cell carcinoma to predict the prognosis and immune infiltration. Front. Immunol. 12, 659184 (2021).
Wieland, A. et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 597, 274–278 (2021).
Xie, M. et al. Immune landscape in molecular subtypes of human papillomavirus-negative head and neck cancer. Mol. Carcinog. 63, 120–135 (2024).
Kurten, C. H. L. et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 12, 7338 (2021).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Sun, Y. et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 184, 404–421 e416 (2021).
de Andrade, L. F., et al. Discovery of specialized NK cell populations infiltrating human melanoma metastases. JCI Insight 4, e133103 (2019).
Larsson, L. C., Anderson, P., Widner, H. & Korsgren, O. Enhanced survival of porcine neural xenografts in mice lacking CD1d1, but no effect of NK1.1 depletion. Cell Transpl. 10, 295–304 (2001).
Kuga, R. et al. HPV infection in squamous cell carcinoma of the hypopharynx, larynx, and oropharynx with multisite involvement. Am. J. Surg. Pathol. 47, 955–966 (2023).
Du, M. et al. Clinicopathologic characteristics of HPV-associated head and neck squamous cell carcinoma in Southern China: long-term retrospective study of 400 cases. Ther. Adv. Med Oncol. 16, 17588359241242962 (2024).
Abdel-Rahman, O. Prognostic value of HPV status among patients with hypopharyngeal carcinoma: a population-based study. Clin. Transl. Oncol. 22, 1645–1650 (2020).
Wu, T. D. et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278 (2020).
Morgan, D. M. et al. Expansion of tumor-reactive CD8(+) T cell clonotypes occurs in the spleen in response to immune checkpoint blockade. Sci. Immunol. 9, eadi3487 (2024).
He, J. et al. Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response. Cell Res. 32, 530–542 (2022).
Tiberti, S. et al. GZMK(high) CD8(+) T effector memory cells are associated with CD15(high) neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome. Nat. Commun. 13, 6752 (2022).
Lan, F. et al. GZMK-expressing CD8(+) T cells promote recurrent airway inflammatory diseases. Nature 638, 490–498 (2025).
Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004 (2018).
Gros, A. et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J. Clin. Investig. 124, 2246–2259 (2014).
Bengsch, B. et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity 48, 1029–1045.e1025 (2018).
Chamoto, K., Yaguchi, T., Tajima, M. & Honjo, T. Insights from a 30-year journey: function, regulation and therapeutic modulation of PD1. Nat. Rev. Immunol. 23, 682–695 (2023).
Poli, A. et al. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 126, 458–465 (2009).
Zwirner, N. W., Domaica, C. I. & Fuertes, M. B. Regulatory functions of NK cells during infections and cancer. J. Leukoc. Biol. 109, 185–194 (2021).
Dogra, P. et al. Tissue determinants of human NK cell development, function, and residence. Cell 180, 749–763 e713 (2020).
Crinier, A. et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity 49, 971–986 e975 (2018).
Khan, M. et al. Transcriptional signature of CD56(bright) NK cells predicts favourable prognosis in bladder cancer. Front. Immunol. 15, 1474652 (2024).
Li, G. et al. TGF-beta-dependent lymphoid tissue residency of stem-like T cells limits response to tumor vaccine. Nat. Commun. 13, 6043 (2022).
Schenkel, J. M. & Pauken, K. E. Localization, tissue biology and T cell state - implications for cancer immunotherapy. Nat. Rev. Immunol. 23, 807–823 (2023).
Eberhardt, C. S. et al. Functional HPV-specific PD-1(+) stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021).
Mogilenko, D. A. et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as a conserved hallmark of inflammaging. Immunity 54, 99–115 e112 (2021).
Jonsson, A. H. et al. Granzyme K(+) CD8 T cells form a core population in inflamed human tissue. Sci. Transl. Med 14, eabo0686 (2022).
Lau, C. M., Wiedemann, G. M. & Sun, J. C. Epigenetic regulation of natural killer cell memory. Immunol. Rev. 305, 90–110 (2022).
Verron, Q., et al. NK cells integrate signals over large areas when building immune synapses, but require local stimuli for degranulation. Sci. Signal. 14, eabe2740 (2021).
Park, S. L. et al. Tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin. Nature 565, 366–371 (2019).
Sun, K. et al. scRNA-seq of gastric tumor shows complex intercellular interaction with an alternative T cell exhaustion trajectory. Nat. Commun. 13, 4943 (2022).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Bottcher, J. P. et al. NK Cells stimulate recruitment of cDC1 into the tumor microenvironment, promoting cancer immune control. Cell 172, 1022–1037 e1014 (2018).
Viel, S. et al. TGF-beta inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal 9, ra19 (2016).
Park, M. H., Lee, J. S. & Yoon, J. H. High expression of CX3CL1 by tumor cells correlates with a good prognosis and increased tumor-infiltrating CD8+ T cells, natural killer cells, and dendritic cells in breast carcinoma. J. Surg. Oncol. 106, 386–392 (2012).
Edge, S. & Byrd, D. (eds) AJCC Cancer Staging Manual 7th edn,(Springer, 2017).
Cai, Z. et al. Single-cell RNA sequencing reveals pro-invasive cancer-associated fibroblasts in hypopharyngeal squamous cell carcinoma. Cell Commun. Signal 21, 292 (2023).
Wang, X. et al. Risk model-guided identification of MTDH expression as a marker for ferroptosis induction therapy in head and neck squamous cell carcinoma. Am. J. Cancer Res. 13, 5236–5253 (2023).
Gao, C., Zhang, M. & Chen, L. The comparison of two single-cell sequencing platforms: bd rhapsody and 10× Genomics Chromium. Curr. Genom. 21, 602–609 (2020).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Zhou, B. & Jin, W. Visualization of single cell RNA-Seq data using t-SNE in R. Methods Mol. Biol. 2117, 159–167 (2020).
Becht, E., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018).
Ji, Z. & Ji, H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 44, e117 (2016).
Yeung, K.Y. & Ruzzo, W.L. Principal component analysis for clustering gene expression data. Bioinformatics 17, 763–774 (2001).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Ju, W. T. et al. Stathmin guides personalized therapy in oral squamous cell carcinoma. Cancer Sci. 111, 1303–1313 (2020).
Li, R. et al. T-cell receptor sequencing reveals hepatocellular carcinoma immune characteristics according to Barcelona Clinic liver cancer stages within liver tissue and peripheral blood. Cancer Sci. 115, 94–108 (2024).
Acknowledgements
This work was supported by the Natural Science Foundation of China (Nos. 82173341 [G.Z.], 82073009 [Y.L.], 82103631 [C.L.], 82303946 [S.L.], 82301286 [L.S.], 82303717 [J.W.]), the National Science and Technology Council, Taiwan (NSTC 113-2639-B-039-001-ASP and T-Star Center NSTC 114-2634-F-039-001 [M.H.]), The Featured Areas Research Center Program by the Ministry of Education (MOE) in Taiwan, The Natural Science Foundation of Hunan (No.2024JJ5558 [G.L.]) and the Fundamental Research Funds for the Central Universities of Central South University (1053320231330 [W.X.]).
Author information
Authors and Affiliations
Contributions
G.L., W.X., H.W., X.Z., M.H., and Y.L. designed the experiments. G.L., H.W., L.S., C.L., X.Z., Y.Q., and Y.L. supervised the clinical work and data. G.L., L.G., W.X., W.W., Y.W., and Y.L. performed the human sample experiments. G.L., W.X., H.Z., J.W., and L.S. performed the mouse experiments. W.X., G.Z., Z.S., J.B., X.X., and X.Y. performed the bioinformation analysis. G.L., C.L., W.X., X.W., D.H., and Y.L. analyzed the results. G.L., X.W., C.L., J.H., N.Z., M.H. and Y.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Dhifaf Sarhan, who co-reviewed with Nerea Almazán, Jian Hou, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, G., Xiao, W., Wu, H. et al. ZNF683+ NK cells govern chemotherapy sensitivity in advanced HPSCC via reshaping immune microenvironment. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68676-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68676-x