Abstract
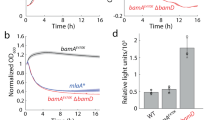
Virtually all integral outer membrane proteins (OMPs) produced by Gram-negative bacteria contain a unique ‘β barrel’ structure that serves as a membrane spanning domain. The universal barrel assembly machine (BAM) catalyzes OMP assembly (folding and membrane insertion) in vivo, and purified Escherichia coli BAM that is reconstituted into proteoliposomes catalyzes OMP assembly in vitro. Here we show that BAM also catalyzes the assembly of OMPs into outer membrane fractions (‘native OMs’) that are purified by optimized conventional methods. Interestingly, we found that OMP assembly was moderately impaired when native OMs were isolated from a mlaA- strain that is deficient in maintaining OM lipid homeostasis but was strongly reduced when native OMs were isolated from a pldA- strain that is deficient in a parallel pathway. We also found that the mlaA and pldA deletions altered the OM phospholipid profile to different degrees that correlated with the degree to which the mutations impaired OMP assembly. Taken together, our results provide direct evidence that the mla and pldA pathways play distinct roles in maintaining OM homeostasis and strongly suggest that OM phospholipids play a more significant role in OMP biogenesis than previously appreciated.
Similar content being viewed by others
Data availability
All of the lipidomics data are provided in Supplementary Data 2. The lipidomics data are also available at figshare (https://doi.org/10.6084/m9.figshare.30780602.v1). The accession codes used in this study are all from the Protein Database (PDB): 6R7L, 1M5Y, 5D0O, 4Q35, 1QD6. 5NUP, 5UWA, 6ZY3, and 3SLJ. Source data are provided with this paper. MS strategies were targeted and signals were analyzed based on known lipid fragmentation conventions. Consequently spectra were not acquired for individual lipids. Method details including instrument parameters and specific MRM’s are included in the source data file. Source data are provided with this paper.
Code availability
The code used in this study is available at Zenodo (https://doi.org/10.5281/zenodo.14755324).
References
De Oliveira, D. M. P. et al. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 33, 10.1128/CMR.00181-19 (2020).
Boucher, H. W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281 (2012).
Luther, A. et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458 (2019).
Imai, Y. et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019).
WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization. (2024).
Silhavy, T. J., Kahne, D. & Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010).
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol. Biol. Rev. 67, 593–656 (2003).
McMorran, L. M., Brockwell, D. J. & Radford, S. E. Mechanistic studies of the biogenesis and folding of outer membrane proteins in vitro and in vivo: what have we learned to date? Arch. Biochem Biophys. 564, 265–280 (2014).
Konovalova, A., Kahne, D. E. & Silhavy, T. J. Outer membrane biogenesis. Annu Rev. Microbiol 71, 539–556 (2017).
Koebnik, R., Locher, K. P. & Van Gelder, P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol 37, 239–253 (2000).
Fairman, J. W., Noinaj, N. & Buchanan, S. K. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 21, 523–531 (2011).
Horne, J. E., Brockwell, D. J. & Radford, S. E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 295, 10340–10367 (2020).
Lauber, F., Deme, J. C., Lea, S. M. & Berks, B. C. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564, 77–82 (2018).
Schalk, I. J., Mislin, G. L. & Brillet, K. Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 69, 37–66 (2012).
Plummer, A. M. & Fleming, K. G. From chaperones to the membrane with a BAM!. Trends Biochem Sci. 41, 872–882 (2016).
Mas, G., Thoma, J. & Hiller, S. The periplasmic chaperones Skp and SurA. Subcell. Biochem 92, 169–186 (2019).
Rouvière, P. E. & Gross, C. A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10, 3170–3182 (1996).
Sklar, J. G., Wu, T., Kahne, D. & Silhavy, T. J. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 (2007).
Vertommen, D., Ruiz, N., Leverrier, P., Silhavy, T. J. & Collet, J. F. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9, 2432–2443 (2009).
Denoncin, K., Schwalm, J., Vertommen, D., Silhavy, T. J. & Collet, J. F. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics 12, 1391–1401 (2012).
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 (2003).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005).
Silale, A. et al. Discovery of a distinct BAM complex in the Bacteroidetes. bioRxiv, 2025.2001.2031.636011 (2025).
Webb, C. T., Heinz, E. & Lithgow, T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 20, 612–620 (2012).
Arnold, T., Zeth, K. & Linke, D. Omp85 from the Thermophilic Cyanobacterium Thermosynechococcus elongatus Differs from Proteobacterial Omp85 in Structure and Domain Composition*. J. Biol. Chem. 285, 18003–18015 (2010).
Malinverni, J. C. et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol 61, 151–164 (2006).
Hart, E. M., Gupta, M., Wühr, M. & Silhavy, T. J. The gain-of-function allele bamA(E470K) bypasses the essential requirement for BamD in β-barrel outer membrane protein assembly. Proc. Natl. Acad. Sci. USA 117, 18737–18743 (2020).
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013).
Doyle, M. T. & Bernstein, H. D. Function of the Omp85 Superfamily Of Outer Membrane Protein Assembly Factors And Polypeptide Transporters. Annu Rev. Microbiol 76, 259–279 (2022).
Doyle, M. T. & Bernstein, H. D. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat. Commun. 10, 3358 (2019).
Wu, R. et al. Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM. Nat. Commun. 12, 7131 (2021).
Doyle, M. T. et al. Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Cell 185, 1143–1156.e1113 (2022).
Tomasek, D. et al. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583, 473–478 (2020).
Shen, C. et al. Structural basis of BAM-mediated outer membrane β-barrel protein assembly. Nature 617, 185–193 (2023).
Rassam, P. et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 523, 333–336 (2015).
Webby, M. N. et al. Lipids mediate supramolecular outer membrane protein assembly in bacteria. Sci. Adv. 8, eadc9566 (2022).
Benn, G. et al. Phase separation in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 118, e2112237118 (2021).
Nyenhuis, D. A. et al. Evidence for the supramolecular organization of a bacterial outer-membrane protein from in vivo pulse electron paramagnetic resonance spectroscopy. J. Am. Chem. Soc. 142, 10715–10722 (2020).
Mamou, G. et al. Peptidoglycan maturation controls outer membrane protein assembly. Nature 606, 953–959 (2022).
Kumar, S. et al. Immobile lipopolysaccharides and outer membrane proteins differentially segregate in growing Escherichia coli. Proc. Natl. Acad. Sci. 122, e2414725122 (2025).
Gunasinghe, S. D. et al. The WD40 protein BamB mediates coupling of bam complexes into assembly precincts in the bacterial outer membrane. Cell Rep. 23, 2782–2794 (2018).
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. 106, 8009–8014 (2009).
Sutterlin, H. A. et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. 113, E1565–E1574 (2016).
Lundstedt, E., Kahne, D. & Ruiz, N. Assembly and Maintenance of Lipids at the Bacterial Outer Membrane. Chem. Rev. 121, 5098–5123 (2021).
May, K. L. & Silhavy, T. J. The Escherichia coli Phospholipase PldA Regulates Outer Membrane Homeostasis via Lipid Signaling. mBio 9, https://doi.org/10.1128/mbio.00379-00318 (2018).
Guest, R. L., Lee, M. J., Wang, W. & Silhavy, T. J. A periplasmic phospholipase that maintains outer membrane lipid asymmetry in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 120, e2302546120 (2023).
Hughes, G. W. et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 4, 1692–1705 (2019).
Tang, X. et al. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat. Struct. Mol. Biol. 28, 81–91 (2021).
Ruiz, N., Davis, R. M. & Kumar, S. YhdP, TamB, and YdbH Are Redundant but Essential for Growth and Lipid Homeostasis of the Gram-Negative Outer Membrane. mBio 12, e0271421 (2021).
Douglass, M. V., McLean, A. B. & Trent, M. S. Absence of YhdP, TamB, and YdbH leads to defects in glycerophospholipid transport and cell morphology in Gram-negative bacteria. PLoS Genet 18, e1010096 (2022).
White, D. A., Lennarz, W. J. & Schnaitman, C. A. Distribution of lipids in the wall and cytoplasmic membrane subfractions of the cell envelope of Escherichia coli. J. Bacteriol. 109, 686–690 (1972).
Lugtenberg, E. J. J. & Peters, R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochimica et. Biophysica Acta (BBA) - Lipids Lipid Metab. 441, 38–47 (1976).
Hussain, S., Peterson, J. H. & Bernstein, H. D. Bam complex-mediated assembly of bacterial outer membrane proteins synthesized in an in vitro translation system. Sci. Rep. 10, 4557 (2020).
Hussain, S. & Bernstein, H. D. The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J. Biol. Chem. 293, 2959–2973 (2018).
Roman-Hernandez, G., Peterson, J. H. & Bernstein, H. D. Reconstitution of bacterial autotransporter assembly using purified components. Elife 3, e04234 (2014).
Hagan, C. L., Kim, S. & Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 (2010).
Norell, D. et al. Versatile in vitro system to study translocation and functional integration of bacterial outer membrane proteins. Nat. Commun. 5, 5396 (2014).
Rath, P. et al. High-throughput screening of BAM inhibitors in native membrane environment. Nat. Commun. 14, 5648 (2023).
Bauman, S. J. & Kuehn, M. J. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 8, 2400–2408 (2006).
Kato, S., Kowashi, Y. & Demuth, D. R. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Micro. Pathog. 32, 1–13 (2002).
Schwechheimer, C. & Kuehn, M. J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol 13, 605–619 (2015).
Kulp, A. & Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev. Microbiol 64, 163–184 (2010).
Walker, M. E. et al. Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition. Nat. Commun. 16, 3395 (2025).
Nikaido, H. & Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 49, 1–32 (1985).
Hussain, S., Peterson, J. H. & Bernstein, H. D. Reconstitution of Bam Complex-Mediated Assembly of a Trimeric Porin into Proteoliposomes. mBio 12, e0169621 (2021).
Peterson, J. H., Yang, L., Gumbart, J. C. & Bernstein, H. D. Conserved lipid-facing basic residues promote the insertion of the porin OmpC into the E. coli outer membrane. mBio 16, e03319–03324 (2025).
Machin, J. M., Kalli, A. C., Ranson, N. A. & Radford, S. E. Protein-lipid charge interactions control the folding of outer membrane proteins into asymmetric membranes. Nat. Chem. (2023).
Nikaido, H. Isolation of outer membranes. Methods Enzymol. 235, 225–234 (1994).
Hobb, R. I., Fields, J. A., Burns, C. M. & Thompson, S. A. Evaluation of procedures for outer membrane isolation from Campylobacter jejuni. Microbiol. (Read.) 155, 979–988 (2009).
Patel, G. J. & Kleinschmidt, J. H. The lipid bilayer-inserted membrane protein bama of escherichia coli facilitates insertion and folding of outer membrane protein a from its complex with skp. Biochemistry 52, 3974–3986 (2013).
Pavlova, O., Peterson, J. H., Ieva, R. & Bernstein, H. D. Mechanistic link between β barrel assembly and the initiation of autotransporter secretion. Proc. Natl. Acad. Sci. USA 110, E938–E947 (2013).
Dautin, N., Barnard, T. J., Anderson, D. E. & Bernstein, H. D. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. Embo j. 26, 1942–1952 (2007).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Krizsan, A. et al. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew. Chem. Int Ed. Engl. 53, 12236–12239 (2014).
Seefeldt, A. C. et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 22, 470–475 (2015).
Freudl, R. et al. An outer membrane protein (OmpA) of Escherichia coli K-12 undergoes a conformational change during export. J. Biol. Chem. 261, 11355–11361 (1986).
Kaur, H. et al. The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature 593, 125–129 (2021).
Ruiz, N., Falcone, B., Kahne, D. & Silhavy, T. J. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005).
Lee, J. et al. Characterization of a stalled complex on the β-barrel assembly machine. Proc. Natl. Acad. Sci. USA 113, 8717–8722 (2016).
Osborn, M. J., Gander, J. E., Parisi, E. & Carson, J. Mechanism of assembly of the outer membrane of salmonella typhimurium: isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247, 3962–3972 (1972).
Berezhnoy, N. V. et al. Transient complexity of e. coli lipidome is explained by fatty acyl synthesis and cyclopropanation. Metabolites 12, 784 (2022).
Allwood, J. W. et al. A workflow for bacterial metabolic fingerprinting and lipid profiling: application to Ciprofloxacin challenged Escherichia coli. Metabolomics 11, 438–453 (2015).
Alves, E. et al. Photodynamic oxidation of Escherichia coli membrane phospholipids: new insights based on lipidomics. Rapid Commun. Mass Spectrom. 27, 2717–2728 (2013).
Oursel, D. et al. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 21, 3229–3233 (2007).
Liu, T., Vora, H. & Khosla, C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab. Eng. 12, 378–386 (2010).
Seurinck, S., Deschepper, E., Deboch, B., Verstraete, W. & Siciliano, S. Characterization of escherichia coli isolates from different fecal sources by means of classification tree analysis of fatty acid methyl ester (fame) profiles. Environ. Monit. Assess. 114, 433–445 (2006).
Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 105, 5537–5542 (2008).
Lundstedt, E. A., Simpson, B. W. & Ruiz, N. LptB-LptF coupling mediates the closure of the substrate-binding cavity in the LptB(2) FGC transporter through a rigid-body mechanism to extract LPS. Mol. Microbiol 114, 200–213 (2020).
Braun, M. & Silhavy, T. J. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol 45, 1289–1302 (2002).
Stanley, A. M., Treubrodt, A. M., Chuawong, P., Hendrickson, T. L. & Fleming, K. G. Lipid chain selectivity by outer membrane phospholipase A. J. Mol. Biol. 366, 461–468 (2007).
Zheng, L., Lin, Y., Lu, S., Zhang, J. & Bogdanov, M. Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim Biophys. Acta Mol. Cell Biol. Lipids 1862, 1404–1413 (2017).
Jouhet, J. Importance of the hexagonal lipid phase in biological membrane organization. Front. Plant Sci. ume 4, 2013 (2013).
Kruijff, B. d. Lipid polymorphism and biomembrane function. Curr. Opin. Chem. Biol. 1, 564–569 (1997).
Marsh, D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 93, 3884–3899 (2007).
Kamischke, C. et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. eLife 8, e40171 (2019).
Cuesta-Seijo, J. A. et al. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure 18, 1210–1219 (2010).
Khan, M. A. & Bishop, R. E. Molecular mechanism for lateral lipid diffusion between the outer membrane external leaflet and a beta-barrel hydrocarbon ruler. Biochemistry 48, 9745–9756 (2009).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Yap, M. N. & Bernstein, H. D. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell 34, 201–211 (2009).
Yap, M. N. & Bernstein, H. D. Mutations in the Escherichia coli ribosomal protein L22 selectively suppress the expression of a secreted bacterial virulence factor. J. Bacteriol. 195, 2991–2999 (2013).
Wang, X., Nyenhuis, S. B. & Bernstein, H. D. The translocation assembly module (TAM) catalyzes the assembly of bacterial outer membrane proteins in vitro. Nat. Commun. 15, 7246 (2024).
Nilaweera, T. D., Nyenhuis, D. A., Nakamoto, R. K. & Cafiso, D. S. Disulfide chaperone knockouts enable in vivo double spin labeling of an outer membrane transporter. Biophys. J. 117, 1476–1484 (2019).
Cian, M. B., Giordano, N. P., Mettlach, J. A., Minor, K. E. & Dalebroux, Z. D. Separation of the Cell Envelope for Gram-negative Bacteria into Inner and Outer Membrane Fractions with Technical Adjustments for Acinetobacter baumannii. J. Vis. Exp., (2020).
Hancock, R. E. W. Hancock Laboratory Methods. Department of Microbiology and Immunology, University of British Columbia, British Columbia, Canada. <https://cmdr.ubc.ca/bobh/method/outer-membrane-preparation-one-step-sucrose-gradient-procedure/> (accessed in 2022).
Garrett, T. A., O’Neill, A. C. & Hopson, M. L. Quantification of cardiolipin molecular species in Escherichia coli lipid extracts using liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 26, 2267–2274 (2012).
Johnson, R. C., Lacroix, I. & Schwarz, B. matrixBP: An R package to generate matrix bar plots (0.1.1). Zenodo., <https://doi.org/10.5281/zenodo.14755324> (2025).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
The PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC.
Kater, L. et al. Partially inserted nascent chain unzips the lateral gate of the Sec translocon. EMBO Rep. 20, EMBR201948191 (2019).
Bitto, E. & McKay, D. B. Crystallographic structure of sura, a molecular chaperone that facilitates folding of outer membrane porins. Structure 10, 1489–1498 (2002).
Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Snijder, H. J. et al. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401, 717–721 (1999).
Abellón-Ruiz, J. et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017).
Ekiert, D. C. et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285.e217 (2017).
Barnard, T. J. et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J. Mol. Biol. 415, 128–142 (2012).
Acknowledgements
We thank Natividad Ruiz (Ohio State University) for providing strain NR698 as well as her unpublished observations, Tom Silhavy (Princeton University) for providing anti-LptD, and Nidhi Kundu (Laboratory of Molecular Biology, NIDDK) for providing technical support and assistance with the TEM imaging. The TEM images were collected at the NIDDK Cryo-EM core facility. We would also like to thank Russell Bishop (University of Toronto) and Zhixin Lyu (Genetics and Biochemistry Branch, NIDDK) for providing insightful comments on the manuscript. This work was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases within the National Institutes of Health (NIH). The contributions of the authors were made as part of their official duties as NIH federal employees, are in compliance with agency policy requirements, and are considered Works of the United States Government. However, the findings and conclusions presented in this paper are those of the authors and do not necessarily reflect the views of the NIH or the U.S. Department of Health and Human Services.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
This study was designed and analyzed by T.D.N. and H.D.B. All of the experimental work, except the lipidomics, was performed by T.D.N. The lipidomics method was developed and lipidomics data was acquired by N.T.B. and B.S. The lipidomic data were analyzed, and corresponding figures were generated by I.S.L., N.T.B., and B.S. The manuscript was written by T.D.N., B.S., and H.D.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nilaweera, T.D., Brandes, N.T., LaCroix, I.S. et al. Phospholipid composition strongly affects the assembly of β barrel proteins into purified bacterial outer membranes. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68743-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68743-3