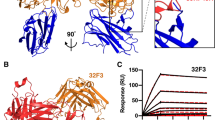
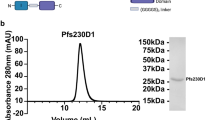
Abstract
Malaria parasite transmission remains a barrier to elimination since asymptomatic individuals sustain the infectious reservoir. Transmission-blocking vaccine (TBV) candidates targeting Plasmodium falciparum (Pf) gametocyte surface proteins Pfs230 and Pfs48/45 have shown promise in clinical trials. Several vaccine candidates have been developed for these antigens, yet it is unclear which elicit the most robust and durable transmission-blocking responses. From structure-function relationships of monoclonal antibodies in complex with both antigens, we report the development of a stabilized tandem antigen chimera (STAC), which presents the most potent epitopes from Pfs230 domain 1 (Pfs230-D1) and Pfs48/45 domain 3 (Pfs48/45-D3) in a single construct, while masking non-functional epitopes using an engineered pseudo-native domain disposition. Iterative structure-guided optimization improved antigen yields and stability, while nanoparticle-based multimerization enhanced the functional transmission-reducing activity elicited by the immunogen in female mice. Immunizations with STAC genetically conjugated to self-assembling protein nanoparticles elicited antibodies with potent transmission-reducing activity comparable or superior to the multimerized Pfs230-D1 and Pfs48/45-D3. These findings establish STAC as a promising next-generation TBV candidate to disrupt malaria transmission and accelerate elimination efforts. More broadly, our results support the engineering of highly ordered and stable multi-domain antigens in a single protein as a strategy for the cost-efficient development of multi-component vaccines.
Similar content being viewed by others
Data availability
The biomolecular structural data generated in this study have been deposited in the RCSB Protein Data Bank under accession codes 9N8N, 9N8I, and 9N8J, and are publicly available as of the date of publication. The cryo-EM volume data has been deposited in the Electron Microscopy Data Bank under entry ID EMD-49130. This paper does not report original code. Raw SMFA data is available in the source data. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon request. Source data are provided with this paper.
References
World Health Organization. World malaria report 2024: addressing inequity in the global malaria response. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2024.
RTS,S. Clinical Trials Partnership. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet Lond. Engl. 386, 31–45 (2015).
Datoo, M. S. et al. Safety and efficacy of malaria vaccine candidate R21/Matrix-M in African children: a multicentre, double-blind, randomised, phase 3 trial. Lancet Lond. Engl. 403, 533–544 (2024).
Dicko, A. et al. Seasonal vaccination with RTS,S/AS01E vaccine with or without seasonal malaria chemoprevention in children up to the age of 5 years in Burkina Faso and Mali: a double-blind, randomised, controlled, phase 3 trial. Lancet Infect. Dis. 24, 75–86 (2024).
Andolina, C. et al. Sources of persistent malaria transmission in a setting with effective malaria control in eastern Uganda: a longitudinal, observational cohort study. Lancet Infect. Dis. 21, 1568–1578 (2021).
Biruksew, A. et al. Schoolchildren with asymptomatic malaria are a potential hotspot for malaria reservoir in Ethiopia: implications for malaria control and elimination efforts. Malar. J. 22, 311 (2023).
Gonçalves, B. P. et al. Examining the human infectious reservoir for Plasmodium falciparum malaria in areas of differing transmission intensity. Nat. Commun. 8, 1133 (2017).
Koepfli, C. et al. Identification of the asymptomatic Plasmodium falciparum and Plasmodium vivax gametocyte reservoir under different transmission intensities. PLoS Negl. Trop. Dis. 15, e0009672 (2021).
Tadesse, F. G. et al. The relative contribution of symptomatic and asymptomatic Plasmodium vivax and Plasmodium falciparum infections to the infectious reservoir in a low-endemic setting in Ethiopia. Clin. Infect. Dis. 66, 1883–1891 (2018).
Penny, M. A., Camponovo, F., Chitnis, N., Smith, T. A. & Tanner, M. Future use-cases of vaccines in malaria control and elimination. Parasite Epidemiol. Control 10, e00145 (2020).
Brady, O. J. et al. Role of mass drug administration in elimination of Plasmodium falciparum malaria: a consensus modelling study. Lancet Glob. Health 5, e680–e687 (2017).
Duffy, P. E., Gorres, J. P., Healy, S. A. & Fried, M. Malaria vaccines: a new era of prevention and control. Nat. Rev. Microbiol. 22, 756–772 (2024).
Yoo, R., Jore, M. M. & Julien, J.-P. Targeting bottlenecks in malaria transmission: antibody-epitope descriptions guide the design of next-generation biomedical interventions. Immunol. Rev. 330, e70001 (2025).
Templeton, T. J. & Kaslow, D. C. Identification of additional members defines a Plasmodium falciparum gene superfamily which includes Pfs48/45 and Pfs230. Mol. Biochem. Parasitol. 101, 223–227 (1999).
Eksi, S. et al. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol. Microbiol. 61, 991–998 (2006).
Marin-Mogollon, C. et al. The Plasmodium falciparum male gametocyte protein P230p, a paralog of P230, is vital for ookinete formation and mosquito transmission. Sci. Rep. 8, 14902 (2018).
van Dijk, M. R. et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell 104, 153–164 (2001).
Bekkering, E. T. et al. Cryo-EM structure of endogenous Pfs230:Pfs48/45 complex with six antibodies reveals mechanisms of malaria transmission-blocking activity. Immunity, Vol. 58, 2899–916.e10 (Elsevier, 2025).
Dietrich, M. H. et al. Cryo-EM structure of endogenous Plasmodium falciparum Pfs230 and Pfs48/45 fertilization complex. Science 389, eady0241 (2025).
MacDonald, N. J. et al. Structural and immunological characterization of recombinant 6-cysteine domains of the Plasmodium falciparum sexual stage protein Pfs230. J. Biol. Chem. 291, 19913–19922 (2016).
Burkhardt, M. et al. Assessment of the impact of manufacturing changes on the physicochemical properties of the recombinant vaccine carrier ExoProtein A. Vaccine 37, 5762–5769 (2019).
Alkema, M. et al. A Pfs48/45-based vaccine to block Plasmodium falciparum transmission: phase 1, open-label, clinical trial. BMC Med. 22, 170 (2024).
Coelho, C. H. et al. Antibody gene features associated with binding and functional activity in malaria vaccine-derived human mAbs. NPJ Vaccines 9, 144 (2024).
Sagara, I. et al. Malaria transmission-blocking vaccines Pfs230D1-EPA and Pfs25-EPA in Alhydrogel in healthy Malian adults; a phase 1, randomised, controlled trial. Lancet Infect. Dis. 23, 1266–1279 (2023).
Healy, S. A. et al. Pfs230 yields higher malaria transmission-blocking vaccine activity than Pfs25 in humans but not mice. J. Clin. Invest. 131, e146221, 146221 (2021).
Coelho, C. H. et al. A human monoclonal antibody blocks malaria transmission and defines a highly conserved neutralizing epitope on gametes. Nat. Commun. 12, 1750 (2021).
Tiono, A. B. et al. A randomized first-in-human phase I trial of differentially adjuvanted Pfs48/45 malaria vaccines in Burkinabé adults. J. Clin. Invest. 134, e175707 (2024).
Naghizadeh, M. et al. Magnitude and durability of ProC6C-AlOH/Matrix-Mtm vaccine-induced malaria transmission-blocking antibodies in Burkinabe adults from a Phase 1 randomized trial. Hum. Vaccines Immunother. 21, 2488075 (2025).
Mulamba, C. et al. Seroprevalence of antibodies to Plasmodium falciparum transmission-blocking target proteins Pfs230D1M and Pfs48/45 in Tanzanian populations of diverse malaria transmission intensity. Front. Immunol. 16, 1589061 (2025).
Bousema, T. et al. The dynamics of naturally acquired immune responses to Plasmodium falciparum sexual stage antigens Pfs230 & Pfs48/45 in a low-endemic area in Tanzania. PloS One 5, e14114 (2010).
Tediosi, F., Maire, N., Penny, M., Studer, A. & Smith, T. A. Simulation of the cost-effectiveness of malaria vaccines. Malar. J. 8, 127 (2009).
Read, D. et al. Transmission-blocking antibodies against multiple, non-variant target epitopes of the Plasmodium falciparum gamete surface antigen Pfs230 are all complement-fixing. Parasite Immunol. 16, 511–519 (1994).
Tang, W. K. et al. A human antibody epitope map of Pfs230D1 derived from analysis of individuals vaccinated with a malaria transmission-blocking vaccine. Immunity 56, 433–443.e5 (2023).
Ivanochko, D. et al. Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230. Immunity 56, 420–432.e7 (2023).
Inklaar, M. R. et al. Pfs230 Domain 7 is targeted by a potent malaria transmission-blocking monoclonal antibody. NPJ Vaccines 8, 186 (2023).
Singh, K. et al. Structure and function of a malaria transmission blocking vaccine targeting Pfs230 and Pfs230-Pfs48/45 proteins. Commun. Biol. 3, 395 (2020).
Fabra-García, A. et al. Highly potent, naturally acquired human monoclonal antibodies against Pfs48/45 block Plasmodium falciparum transmission to mosquitoes. Immunity 56, 406–419.e7 (2023).
Lennartz, F. et al. Structural basis for recognition of the malaria vaccine candidate Pfs48/45 by a transmission blocking antibody. Nat. Commun. 9, 3822 (2018).
Singh, S. K. et al. Pfs230 and Pfs48/45 fusion proteins elicit strong transmission-blocking antibody responses against Plasmodium falciparum. Front. Immunol. 10, 1256 (2019).
Singh, S. K. et al. Preclinical development of a Pfs230-Pfs48/45 chimeric malaria transmission-blocking vaccine. NPJ Vaccines 6, 120 (2021).
Plieskatt, J. et al. ProC6C, a novel multi-stage malaria vaccine, elicits functional antibodies against the minor and central repeats of the Circumsporozoite Protein in human adults. Front. Immunol. 15, 1481829 (2024).
McLeod, B. et al. Vaccination with a structure-based stabilized version of malarial antigen Pfs48/45 elicits ultra-potent transmission-blocking antibody responses. Immunity 55, 1680–1692.e8 (2022).
Dickey, T. H. et al. Design of a stabilized non-glycosylated Pfs48/45 antigen enables a potent malaria transmission-blocking nanoparticle vaccine. NPJ Vaccines 8, 20 (2023).
Dauparas, J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
He, X. et al. A potent cancer vaccine adjuvant system for particleization of short, synthetic CD8+ T cell epitopes. ACS Nano 15, 4357–4371 (2021).
Lee, S.-M. et al. The Pfs230 N-terminal fragment, Pfs230D1+: expression and characterization of a potential malaria transmission-blocking vaccine candidate. Malar. J. 18, 356 (2019).
Miura, K. et al. Qualification of standard membrane-feeding assay with Plasmodium falciparum malaria and potential improvements for future assays. PLoS ONE 8, e57909 (2013).
Kucharska, I. et al. Structural elucidation of full-length Pfs48/45 in complex with potent monoclonal antibodies isolated from a naturally exposed individual. Nat. Struct. Mol. Biol. 32, 1396–1407 (2025).
Xu, J. & Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010).
Lee, S.-M. et al. Expression and purification optimization of an N-terminal Pfs230 transmission-blocking vaccine candidate. Protein Expr. Purif. 160, 56–65 (2019).
Kelly, H. G., Kent, S. J. & Wheatley, A. K. Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Rev. Vaccines 18, 269–280 (2019).
Zhao, L. et al. Nanoparticle vaccines. Vaccine 32, 327–337 (2014).
Wargacki, A. J. et al. Complete and cooperative in vitro assembly of computationally designed self-assembling protein nanomaterials. Nat. Commun. 12, 883 (2021).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013).
Ludwig, J. et al. Glycosylated nanoparticle-based PfCSP vaccine confers long-lasting antibody responses and sterile protection in the mouse malaria model. NPJ Vaccines 8, 52 (2023).
Thera, M. A. et al. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 365, 1004–1013 (2011).
Neafsey, D. E. et al. Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. N. Engl. J. Med. 373, 2025–2037 (2015).
Kiyuka, P. K., Meri, S. & Khattab, A. Complement in malaria: immune evasion strategies and role in protective immunity. FEBS Lett. 594, 2502–2517 (2020).
Eggink, D., Goff, P. H. & Palese, P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 88, 699–704 (2014).
Cardozo, T. et al. Vaccine focusing on cross-subtype HIV-1 gp120 variable loop epitopes. Vaccine 32, 4916–4924 (2014).
Wang, H. et al. Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses. J. Biol. Chem. 293, 830–846 (2018).
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013).
Ringe, R. P. et al. Closing and opening holes in the glycan shield of hiv-1 envelope glycoprotein SOSIP trimers can redirect the neutralizing antibody response to the newly unmasked epitopes. J. Virol. 93, e01656-18 (2019).
Seabright, G. E., Doores, K. J., Burton, D. R. & Crispin, M. Protein and glycan mimicry in HIV vaccine design. J. Mol. Biol. 431, 2223–2247 (2019).
Walls, A. C. et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020).
Huang, H.-Y. et al. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci. Transl. Med. 14, eabm0899 (2022).
Boyington, J. C. et al. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PloS One 11, e0159709 (2016).
Swanson, K. A. et al. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci. Immunol. 5, eaba6466 (2020).
Shrock, E. L. et al. Germline-encoded amino acid-binding motifs drive immunodominant public antibody responses. Science 380, eadc9498 (2023).
Aung, A. et al. Low protease activity in B cell follicles promotes retention of intact antigens after immunization. Science 379, eabn8934 (2023).
Xu, D. et al. Vaccine design via antigen reorientation. Nat. Chem. Biol. 20, 1012–1021 (2024).
Hendricks, G. G. et al. Computationally designed mRNA-launched protein nanoparticle immunogens elicit protective antibody and T cell responses in mice. Sci. Transl. Med. 17, eadu2085 (2025).
Scaria, P. V. et al. mRNA vaccines expressing malaria transmission-blocking antigens Pfs25 and Pfs230D1 induce a functional immune response. NPJ Vaccines 9, 9 (2024).
Kunkeaw, N. et al. A Pvs25 mRNA vaccine induces complete and durable transmission-blocking immunity to Plasmodium vivax. NPJ Vaccines 8, 187 (2023).
Mallory, K. L. et al. Messenger RNA expressing PfCSP induces functional, protective immune responses against malaria in mice. NPJ Vaccines 6, 84 (2021).
Fotoran, W. L. et al. Establishment of an antiplasmodial vaccine based on PfRH5-encoding rna replicons stabilized by cationic liposomes. Pharmaceutics 15, 1223 (2023).
Ganley, M. et al. mRNA vaccine against malaria tailored for liver-resident memory T cells. Nat. Immunol. 24, 1487–1498 (2023).
Ponnudurai, T. et al. Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology 98, 165–173 (1989).
Ramjith, J. et al. Quantifying reductions in Plasmodium falciparum infectivity to mosquitos: a sample size calculator to inform clinical trials on transmission-reducing interventions. Front. Immunol. 13, 899615 (2022).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Marr, C. R., Benlekbir, S. & Rubinstein, J. L. Fabrication of carbon films with ∼500 nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014).
Guo, H. et al. Electron-event representation data enable efficient cryoEM file storage with full preservation of spatial and temporal resolution. IUCrJ 7, 860–869 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. Publ. Protein Soc. 27, 14–25 (2018).
Manalastas-Cantos, K. et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Svergun, D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Acknowledgments
We would like to thank Nicholas Proellochs, Wouter Graumans, Marga van de Vegte-Bolmer, Laura Pelser-Posthumus, Astrid Pouwelsen, Jacqueline Kuhnen and Jolanda Klaassen for assistance with parasite culture and mosquito infections; Samir Benlekbir and Zhijie Li from the SickKids Nanoscale Biomedical Imaging Facility for assistance and insights during cryo-EM data collection; Greg Wasney and James Magnus Jorgensen at the Structural & Biophysical Core (SBC) Facility for assistance. X-ray diffraction experiments for biomolecular crystallography were performed at GM/CA@APS, which has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16 M detector was funded by an NIH–Office of Research Infrastructure Programs High-End Instrumentation grant (1S10OD012289-01A1). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357. X-ray diffraction experiments were also performed at beamline CMCF-ID at the Canadian Light Source, a national research facility of the University of Saskatchewan, which is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. The small-angle X-ray scattering instrument was accessed at the Structural and Biophysical Core Facility, The Hospital for Sick Children, and EM data was collected at the Nanoscale Biomedical Imaging Facility, The Hospital for Sick Children, supported by the Canada Foundation for Innovation and Ontario Research Fund. This work was supported by a National Institutes of Health grant (1R01AI148557-01A1 to J.F.L., R.S.M., and J-P.J.); Bill & Melinda Gates Foundation grant (OPP1156262 to N.P.K. and J-P.J.); a Canadian Institutes of Health Research Project grant (428410 to J-P.J.); and, in part, thanks to funding from the Canada Research Chair program (J-P.J.). M.M.J. is supported by the Netherlands Organization for Scientific Research (Vidi fellowship NWO project number 192.061). S.H. is supported by a Canada Graduate Scholarship - Doctoral. This work was also supported by the Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID).
Author information
Authors and Affiliations
Contributions
Experimental design was collaborative between all co-authors. Experiments were conducted by D.I., K.M., S.H., R.R., Y.S., W-C.H., R.S., K.T., G-J.v.G., E.M.L., S.C., C.M., A.S., and C.S. The manuscript was written by D.I. and J-P.J., and edited by all co-authors. Funding was secured by R.S.M., C.A.L., M.M.J., N.P.K., J.F.L., and J-P.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. Patent applications have been filed that relate to this work.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ivanochko, D., Miura, K., Hailemariam, S. et al. A stabilized tandem antigen chimera that elicits potent malaria transmission-reducing activity. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68761-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68761-1