Abstract
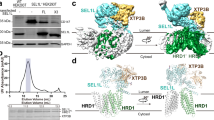
The SEL1L-HRD1 complex represents the most conserved branch of endoplasmic reticulum (ER)-associated degradation (ERAD), a critical quality-control pathway that clears misfolded ER proteins. However, the molecular organization and pathogenic mechanisms of mammalian ERAD have remained elusive. Here, we report the cryo-EM structure of the core mammalian ERAD complex, comprising the ER lectin OS9, SEL1L, and the E3 ubiquitin ligase HRD1. The structure, validated by mutagenesis and crosslinking assays, reveals a dimeric assembly of the core complex in which SEL1L and OS9 form a claw-like configuration in the ER lumen that mediates substrate engagement, while HRD1 dimerizes within the membrane that may facilitate substrate translocation. Furthermore, pathogenic SEL1L mutations at the SEL1L-OS9 (Gly585Asp) and SEL1L-HRD1 (Ser658Pro) interfaces disrupt complex formation and impair ERAD activity. A newly identified disease-associated HRD1 variant (Ala91Asp), located in transmembrane helix 3, impairs HRD1 dimerization and substrate processing. These findings provide structural and functional insights for mammalian SEL1L-HRD1 ERAD and elucidate how mutations destabilizing this machinery contribute to human disease.
Similar content being viewed by others
Data availability
The materials and reagents used are either commercially available or available upon the request. Proteomic data are available in a public database PRIDE (PXD043674 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD043674) and PXD041882). The cryo-EM map and the corresponding atomic model for the ERAD complex structure have been deposited under the accession codes EMD70448 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-70448) (consensus map), EMD-70452 (locally refined map) and 9OG0. All data related to this study are available in the main text or the supplementary material. Source data are provided with this paper.
References
Kanapin, A. et al. Mouse proteome analysis. Genome Res 13, 1335–1344 (2003).
Benham, A. M. Protein secretion and the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4, a012872 (2012).
Lippincott-Schwartz, J., Bonifacino, J. S., Yuan, L. C. & Klausner, R. D. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell 54, 209–220 (1988).
Sommer, T. & Jentsch, S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365, 176–179 (1993).
Jensen, T. J. et al. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129–135 (1995).
Ward, C. L., Omura, S. & Kopito, R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 (1995).
McCracken, A. A. & Brodsky, J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298 (1996).
Hampton, R. Y., Gardner, R. G. & Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7, 2029–2044 (1996).
Bordallo, J., Plemper, R. K., Finger, A. & Wolf, D. H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9, 209–222 (1998).
Biunno, I. et al. Isolation of a pancreas-specific gene located on human chromosome 14q31: expression analysis in human pancreatic ductal carcinomas. Genomics 46, 284–286 (1997).
Kaneko, M., Ishiguro, M., Niinuma, Y., Uesugi, M. & Nomura, Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532, 147–152 (2002).
Hwang, J. & Qi, L. Quality control in the endoplasmic reticulum: crosstalk between ERAD and UPR pathways. Trends Biochem Sci. 43, 593–605 (2018).
Wang, H. H. et al. Hypomorphic variants of SEL1L-HRD1 ER-associated degradation are associated with neurodevelopmental disorders. J. Clin. Invest. https://doi.org/10.1172/JCI170054 (2024).
Weis, D. et al. Biallelic Cys141Tyr variant of SEL1L is associated with neurodevelopmental disorders, agammaglobulinemia, and premature death. J. Clin. Invest. https://doi.org/10.1172/JCI170882 (2024).
Wang, H. H., Biunno, I., Sun, S. & Qi, L. SEL1L-HRD1-mediated ERAD in mammals. Nat. Cell Biol. https://doi.org/10.1038/s41556-025-01690-1 (2025).
Qi, L., Tsai, B. & Arvan, P. New insights into the physiological role of endoplasmic reticulum-associated degradation. Trends Cell Biol. 27, 430–440 (2017).
Bhattacharya, A. & Qi, L. ER-associated degradation in health and disease - from substrate to organism. J. Cell Sci. 132, jcs232850 (2019).
Sha, H. et al. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab. 20, 458–470 (2014).
Sun, S. et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc. Natl. Acad. Sci. USA 111, E582–E591 (2014).
Francisco, A. B. et al. Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J. Biol. Chem. 285, 13694–13703 (2010).
Fujita, H. et al. The E3 ligase synoviolin controls body weight and mitochondrial biogenesis through negative regulation of PGC-1beta. EMBO J. 34, 1042–1055 (2015).
Yagishita, N. et al. Essential role of synoviolin in embryogenesis. J. Biol. Chem. 280, 7909–7916 (2005).
Ji, Y. et al. The Sel1L-Hrd1 endoplasmic reticulum-associated degradation complex manages a key checkpoint in B cell development. Cell Rep. 16, 2630–2640 (2016).
Sun, S. et al. Epithelial Sel1L is required for the maintenance of intestinal homeostasis. Mol. Biol. Cell 27, 483–490 (2016).
Bhattacharya, A. et al. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. EMBO J. 37, e99277 (2018).
Shrestha, N. et al. Sel1L-Hrd1 ER-associated degradation maintains β cell identity via TGFβ signaling. J. Clin. Invest 130, 3499–3510 (2020).
Yoshida, S. et al. Endoplasmic reticulum-associated degradation is required for nephrin maturation and kidney glomerular filtration function. J. Clin. Invest. https://doi.org/10.1172/JCI143988 (2021).
Abdon, B. et al. Muscle specific ER-associated degradation maintains postnatal muscle hypertrophy and systemic energy metabolism. JCI Insight 8, e170387 (2023).
Ji, Y. et al. SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool. Nat. Cell Biol. https://doi.org/10.1038/s41556-023-01138-4 (2023).
Thepsuwan, P. et al. Hepatic SEL1L-HRD1 ER-associated degradation regulates systemic iron homeostasis via ceruloplasmin. Proc. Natl. Acad. Sci. USA 120, e2212644120 (2023).
Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A. & Hampton, R. Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3, 24–29 (2001).
Gardner, R. G. et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control Hrd1p Hrd3p. J. Cell Biol. 151, 69–82 (2000).
Mueller, B., Lilley, B. N. & Ploegh, H. L. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175, 261–270 (2006).
Mueller, B., Klemm, E. J., Spooner, E., Claessen, J. H. & Ploegh, H. L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA 105, 12325–12330 (2008).
Bhamidipati, A., Denic, V., Quan, E. M. & Weissman, J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell 19, 741–751 (2005).
Kim, W., Spear, E. D. & Ng, D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell 19, 753–764 (2005).
Szathmary, R., Bielmann, R., Nita-Lazar, M., Burda, P. & Jakob, C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell 19, 765–775 (2005).
Christianson, J. C., Shaler, T. A., Tyler, R. E. & Kopito, R. R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 (2008).
Hosokawa, N. et al. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914–20924 (2008).
Lilley, B. N. & Ploegh, H. L. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 102, 14296–14301 (2005).
Ye, Y., Shibata, Y., Yun, C., Ron, D. & Rapoport, T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 (2004).
Ye, Y. et al. Inaugural Article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 102, 14132–14138 (2005).
Carvalho, P., Goder, V. & Rapoport, T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 (2006).
Schoebel, S. et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548, 352–355 (2017).
Wu, X. et al. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368, https://doi.org/10.1126/science.aaz2449 (2020).
Hwang, J. et al. Characterization of protein complexes of the endoplasmic reticulum-associated degradation E3 ubiquitin ligase Hrd1. J. Biol. Chem. 292, 9104–9116 (2017).
Huang, C. H., Hsiao, H. T., Chu, Y. R., Ye, Y. & Chen, X. Derlin2 protein facilitates HRD1-mediated retro-translocation of sonic hedgehog at the endoplasmic reticulum. J. Biol. Chem. 288, 25330–25339 (2013).
Huang, C. H., Chu, Y. R., Ye, Y. & Chen, X. Role of HERP and a HERP-related protein in HRD1-dependent protein degradation at the endoplasmic reticulum. J. Biol. Chem. 289, 4444–4454 (2014).
Lin, L. L. et al. SEL1L-HRD1 interaction is required to form a functional HRD1 ERAD complex. Nat. Commun. 15, 1440 (2024).
van der Goot, A. T., Pearce, M. M. P., Leto, D. E., Shaler, T. A. & Kopito, R. R. Redundant and Antagonistic Roles of XTP3B and OS9 in Decoding Glycan and Non-glycan Degrons in ER-Associated Degradation. Mol. Cell 70, 516–530.e516 (2018).
Fujimori, T., Kamiya, Y., Nagata, K., Kato, K. & Hosokawa, N. Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded alpha1-antitrypsin variant. FEBS J. 280, 1563–1575 (2013).
Schulz, J. et al. Conserved cytoplasmic domains promote Hrd1 ubiquitin ligase complex formation for ER-associated degradation (ERAD). J. Cell Sci. 130, 3322–3335 (2017).
Shi, G. et al. ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis. J. Clin. Invest 127, 3897–3912 (2017).
Friberg, M. A., Spiess, M. & Rutishauser, J. Degradation of wild-type vasopressin precursor and pathogenic mutants by the proteasome. J. Biol. Chem. 279, 19441–19447 (2004).
Kikkert, M. et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 279, 3525–3534 (2004).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Pisa, R. & Rapoport, T. A. Disulfide-crosslink analysis of the ubiquitin ligase Hrd1 complex during endoplasmic reticulum-associated protein degradation. J. Biol. Chem. 298, 102373 (2022).
Hwang, C., Sinskey, A. J. & Lodish, H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 (1992).
Kim, G. H. et al. Hypothalamic ER-associated degradation regulates POMC maturation, feeding, and age-associated obesity. J. Clin. Invest 128, 1125–1140 (2018).
Pleiner, T. et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science 369, 433–436 (2020).
Bai, L., You, Q., Feng, X., Kovach, A. & Li, H. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 584, 475–478 (2020).
Jomaa, A. et al. Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 8, 15470 (2017).
Jomaa, A. et al. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 36, 109350 (2021).
Guo, L. et al. Structural insights into the human HRD1 ubiquitin ligase complex. Nat. Commun. 16, 6007 (2025).
Rao, B. et al. The cryo-EM structure of the human ERAD retrotranslocation complex. Sci. Adv. 9, eadi5656 (2023).
Wang, Q. et al. Cryo-EM structure of the human Derlin-1/p97 complex reveals a hexameric channel in ERAD. Commun. Biol. 8, 1481 (2025).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Zhou, Z. et al. Endoplasmic reticulum-associated degradation regulates mitochondrial dynamics in brown adipocytes. Science 368, 54–60 (2020).
Kyostila, K. et al. A SEL1L mutation links a canine progressive early-onset cerebellar ataxia to the endoplasmic reticulum-associated protein degradation (ERAD) machinery. PLoS Genet 8, e1002759 (2012).
Acknowledgements
We thank the Qi, Jomaa, Sun labs (UVA), Arvan lab (UM), Drs. David Castle (UVA), Qinli Hu (UT Southwestern Medical Center), Maciej Gluc (UVA), Ruoya Ho (UVA), and Huan Bao (UVA) for technical support and insightful discussions; Drs. Michael Purdy and David Cooper (MEMC core, UVA), Haoran Yuan (UVA) and Riley Loyd (UVA) for assisting with the cryo-EM data collection and computational support; Dr. Chih-Chi Andrew Hu (Houston Methodist Hospital) for the Derlin-1 and Derlin-2 antibodies; and Drs. Fowzan Alkuraya and Khadijah Bakur for providing the information on the HRD1 A91D variant from the Lifera Omics Database. Cryo-EM data collection was conducted at the Molecular Electron Microscopy Core Facility (MEMC, RRID:SCR_019031) at the University of Virginia (UVA) School of Medicine (NIH G20-RR31199). We thank the Proteomics Resource Facility at University of Michigan Medical School for assisting proteomic assays. This work was supported by The Owens Family Foundation, the Searle Scholars Program (SSP-2023-106), American Cancer Society grant (#134088-IRG-19-143-33-IRG), and NIH 1R35 GM160490 to A.J.; NIH R01DK120047, R01DK120330, R35GM130292 to L.Q. L.L.L. was supported in part by National Ataxia Foundation Post-doctoral Fellowships NAF 918037. E.M. is supported by the cellular and molecular biology training program at UVA through NIH T32GM139787-3. L.E.Z. is supported by the American Heart Association Pre-doctoral Fellowship 25PRE1375196.
Author information
Authors and Affiliations
Contributions
This study was conceived by L.L.L., A.J. and L.Q. L.L.L. purified the ERAD core complex and collected cryo-EM data. E.M. processed cryo-EM data and built the atomic model. L.L.L. designed and performed all the biochemical experiments. L.E.Z. conducted the site directed point mutagenesis and plasmid preps. L.L.L. and L.Q. wrote the manuscript. All authors contributed to data analysis and final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Peer review
Peer review information
Nature Communications thanks Kenji Inaba and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, L.L., Maldosevic, E., Zhou, L.E. et al. Structural basis and pathological implications of the dimeric OS9-SEL1L-HRD1 ERAD Core Complex. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68777-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68777-7