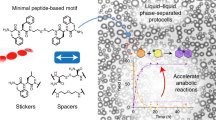
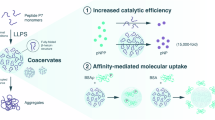
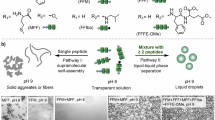
Abstract
The mechanisms governing pH regulation in membrane-less organelles (MLOs) are essentially different from those relying on membrane proteins, yet it remains poorly understood due to the difficulty in directly controlling the conditions across the MLOs interface. Here, we develop a coacervate-based, in vitro model to investigate how liquid-liquid phase separation (LLPS) could contribute to pH regulation in MLOs. We construct peptide-based coacervate droplets using microfluidics and find that charged polymers within the coacervates help create uneven H+/OH- distributions, resulting in pH-regionalized microenvironments similar to the nucleolus. More interestingly, such a pH difference weakens or even disappears following the destruction of LLPS, a phenomenon observed in both nucleolus and coacervate droplets. Based on these findings, we demonstrate the ability to finely tune the local pH of the coacervate droplets over a wide range by incorporating enzymes, which can drive and control cascade reactions, and perform basic molecular biology operations such as polymerase chain reaction (PCR) and in vitro transcription and translation reaction (IVTT). This study highlights the role of LLPS in pH modulation within MLOs and provides insights into the potential of coacervates as protocells for broader applications.
Similar content being viewed by others
Data availability
Unless otherwise stated, all data supporting the results of this study can be found in the article, supplementary, and source data files. Source data are provided with this paper.
Code availability
The molecular dynamics simulation codes supporting the findings of this study are available in the source data.
References
Li, X., Egervari, G., Wang, Y., Berger, S. L. & Lu, Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. 19, 563–578 (2018).
Ravindran, R. et al. Peroxisome biogenesis initiated by protein phase separation. Nature 617, 608–615 (2023).
Stegen, S. & Carmeliet, G. Metabolic regulation of skeletal cell fate and function. Nat. Rev. Endocrinol. 20, 399–413 (2024).
Baker-Austin, C. & Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15, 165–171 (2007).
King, M. R. et al. Macromolecular condensation organizes nucleolar sub-phases to set up a pH gradient. Cell 187, 1889–1906.e1824 (2024).
Dai, Y. et al. Biomolecular condensates regulate cellular electrochemical equilibria. Cell 187, 5951–5966 e5918 (2024).
Lenne, P.-F. & Trivedi, V. Sculpting tissues by phase transitions. Nat. Commun. 13, 664 (2022).
Cook, A. B., Novosedlik, S. & van Hest, J. C. M. Complex Coacervate Materials as Artificial Cells. Acc. Mater. Res. 4, 287–298 (2023).
Wang, Y. et al. Biomolecular condensates mediate bending and scission of endosome membranes. Nature 634, 1204–1210 (2024).
Galvanetto, N. et al. Extreme dynamics in a biomolecular condensate. Nature 619, 876–883 (2023).
Xu, C., Martin, N., Li, M. & Mann, S. Living material assembly of bacteriogenic protocells. Nature 609, 1029–1037 (2022).
Jambon-Puillet, E. et al. Phase-separated droplets swim to their dissolution. Nat. Commun. 15, 3919 (2024).
Testa, A. et al. Sustained enzymatic activity and flow in crowded protein droplets. Nat. Commun. 12, 6293 (2021).
Wang, Z. et al. Coacervate Microdroplets as Synthetic Protocells for Cell Mimicking and Signaling Communications. Small Methods 7, e2300042 (2023).
Chen, H., Guo, J., Bian, F. & Zhao, Y. Microfluidic technologies for cell deformability cytometry. Smart Med 1, e20220001 (2022).
Shang, L., Cheng, Y. & Zhao, Y. Emerging Droplet Microfluidics. Chem. Rev. 117, 7964–8040 (2017).
Yuan, Y. et al. Digital droplet RT-LAMP increases speed of SARS-CoV-2 viral RNA detection. Smart Med. 3, e20240008 (2024).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2020).
Tafech, A., Beaujean, C., Usson, Y. & Stéphanou, A. Generalization of the Ratiometric Method to Extend pH Range Measurements of the BCECF Probe. Biomolecules 13, 442 (2023).
Aryan, F. et al. Nucleolus activity-dependent recruitment and biomolecular condensation by pH sensing. Mol. Cell 83, 4413–4423.e4410 (2023).
Mars, J. C. et al. The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. NAR Cancer 2, zcaa032 (2020).
Choi, S., Meyer, M. O., Bevilacqua, P. C. & Keating, C. D. Phase-specific RNA accumulation and duplex thermodynamics in multiphase coacervate models for membraneless organelles. Nat. Chem. 14, 1110–1117 (2022).
Souza, P. C. T. et al. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021).
Liu, Y., Wang, X., Wan, Z., Ngai, T. & Tse, Y. S. Capturing coacervate formation and protein partition by molecular dynamics simulation. Chem. Sci. 14, 1168–1175 (2023).
Wang, C., Yang, X., Wang, Q., Zhang, L. & Shang, L. Glucose Responsive Coacervate Protocells from Microfluidics for Diabetic Wound Healing. Adv. Sci. 11, 2400712 (2024).
Onosakponome Iruoghene, E. A. L., Eze, S. O., Onyebuchi & Chilaka, F. Chiemeka Kinetics and Thermodynamic Properties of Glucose Oxidase Obtained from Aspergillus fumigatus ASF4. Trop. J. Nat. Prod. Res. 6, 438–445 (2022).
Cesareo, S. D. & Langton, S. R. Kinetic properties of Helicobacter pylori urease compared with jack bean urease. FEMS Microbiol. Lett. 78, 15–21 (1992).
Gao, H., Kong, J., Li, Z., Xiao, G. & Meng, X. Quantitative analysis of temperature, salinity and pH on WSSV proliferation in Chinese shrimp Fenneropenaeus chinensis by real-time PCR. Aquaculture 312, 26–31 (2011).
Tsanai, M., Frederix, P. W. J. M., Schroer, C. F. E., Souza, P. C. T. & Marrink, S. J. Coacervate formation studied by explicit solvent coarse-grain molecular dynamics with the Martini model. Chem. Sci. 12, 8521–8530 (2021).
Guo, Y., Huang, S.-F., Mabuchi, T. & Tokumasu, T. Analysis of structural and water diffusional properties of ionomer thin film by coarse-grained molecular dynamics simulation. J. Mol. Liq. 391, 123190 (2023).
Al-Jubair, T. et al. Characterization of human aquaporin protein-protein interactions using microscale thermophoresis (MST). STAR Protoc. 3, 101316 (2022).
Jerabek-Willemsen, M., Wienken, C. J., Braun, D., Baaske, P. & Duhr, S. Molecular Interaction Studies Using Microscale Thermophoresis. ASSAY Drug Dev. Technol. 9, 342–353 (2011).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2024YFA0919100), the National Natural Science Foundation of China (32271383), and the Shenzhen Medical Research Fund (B2401006). We are particularly thankful to the Core Facility of Shanghai Medical College, Fudan University.
Author information
Authors and Affiliations
Contributions
C.W. designed and performed the majority of the experiments, analyzed most of the data, and wrote the manuscript. Z.F. contributed to the analysis of turbidity data. L.Z. carried out part of the cell experiments. L.S. supervised the project, contributed to the experimental design, and co-wrote the manuscript. Y.Z. contributed to the experimental design and manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Dhwanit Dave and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, C., Fang, Z., Zhang, L. et al. Coacervate droplets as pH-regionalized protocells. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68980-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68980-6