Abstract
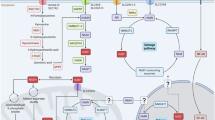
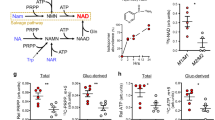
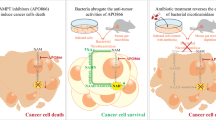
Nicotinamide adenine dinucleotide (NAD) is synthesized through both amidated salvage and deamidated pathways. Although NAD-producing enzymes are often overexpressed in cancer cells to meet the high metabolic demands of rapid proliferation and are considered oncogenic, we report that physiological levels of nicotinic acid phosphoribosyl transferase (NAPRT), the first enzyme in the Preiss-Handler arm of the deamidated pathways, suppress tumorigenesis. We show that NAPRT is enriched in gut epithelial cells, where it sustains the NAD pool for an efficient response to stress-induced acute NAD depletion. Consequently, NAPRT deficiency impairs the activity of poly-(ADP-ribose) polymerases and DNA repair, sensitizes mice to chemical-induced colitis and tumorigenesis, as well as to age-associated spontaneous tumor development. Moreover, low NAPRT expression correlates with poor prognosis in several human cancer types. Thus, homeostatic levels of deamidated NAD biosynthesis contribute to tumor suppression, and boosting this pathway may offer a strategy for cancer prevention.
Similar content being viewed by others
Data availability
The RNA-seq and scRNA-seq datasets of this study have been deposited to Gene Expression Omnibus under the following accession numbers: RNA-seq dataset of the AOM experiment: GSE271834 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE271834]RNA-seq dataset of the DSS-colitis experiment: GSE271250 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE271250]. scRNA-seq datasets of colon tissues: GSE271836 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE271836]
scRNA-seq datasets of small intestinal tissues: GSE261216 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE261216], Metabolomics data of this study have been deposited to massive.ucsd.edu with the accession number MSV000100103, https://doi.org/10.25345/C56M33H4M. The detailed data of 277 identified or annotated metabolites are provided in Supplementary Data 1. LC-MS data for targeted measurement of mouse tissue NAD metabolites have been deposited to massive.ucsd.edu with the accession number MSV000100196, https://doi.org/10.25345/C5639KJ8W. The representative spectral peaks are provided in Supplementary Data 8. LC-MS parameters and representative spectral peaks from targeted measurement of NUA in mouse kidney are provided in Supplementary Data 9. The LC-MS raw data files for NAD metabolites in mouse plasma have been uploaded to metabolomicsworkbench.org under tracking ID 6868 (NA and NAM) and 6875 (NAR), https://doi.org/10.21228/M8BN9Z. The LC-MS data and representative spectral peaks are provided in Supplementary Data 10. Public datasets used in this study are: GSE111889. GSE201348. HTAN VUMC [https://cellxgene.cziscience.com/collections/a48f5033-3438-4550-8574-cdff3263fdfd]. TCGA PanCancer datasets: https://www.cbioportal.org/. Source data containing the exact p-values for all statistical tests are provided with this paper.
Code availability
Scripts used for scRNA-seq analyses in this study are available with no restriction at github: https:github.com/JohnXu24/NAPRTKO_scRNA. Published software packages and pipelines are used for RNA-seq and scRNA-seq analyses. Detailed description of these data analyses is included in Methods.
References
Houtkooper, R. H., Canto, C., Wanders, R. J. & Auwerx, J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194–223 (2010).
Cantó, C., Menzies, K. J. & Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 22, 31–53 (2015).
Bogan, K. L. & Brenner, C. Nicotinic Acid, Nicotinamide, and Nicotinamide Riboside: A Molecular Evaluation of NAD + Precursor Vitamins in Human Nutrition. Annu. Rev. Nutr. 28, 115–130 (2008).
Grozio, A. et al. Slc12a8 is a nicotinamide mononucleotide transporter. Nat. Metab. 1, 47–57 (2019).
Yoshino, J., Baur, J. A. & Imai, S. I. NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 27, 513–528 (2018).
Chini, C. C. S., Zeidler, J. D., Kashyap, S., Warner, G. & Chini, E. N. Evolving concepts in NAD(+) metabolism. Cell Metab. 33, 1076–1087 (2021).
Covarrubias, A. J., Perrone, R., Grozio, A. & Verdin, E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 22, 119–141 (2021).
Fang, E. F. et al. NAD(+) in Aging: Molecular Mechanisms and Translational Implications. Trends Mol. Med 23, 899–916 (2017).
Chowdhry, S. et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569, 570–575 (2019).
Espindola-Netto, J. M. et al. Preclinical efficacy of the novel competitive NAMPT inhibitor STF-118804 in pancreatic cancer. Oncotarget 8, 85054–85067 (2017).
Moschen, A. R. et al. Visfatin, an adipocytokine with proinflammatory and immunomodulating properties. J. Immunol. 178, 1748–1758 (2007).
Gerner, R. R. et al. NAD metabolism fuels human and mouse intestinal inflammation. Gut 67, 1813–1823 (2018).
Baixauli, F. et al. Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses. Cell Metab. 22, 485–498 (2015).
Cameron, A. M. et al. Inflammatory macrophage dependence on NAD(+) salvage is a consequence of reactive oxygen species-mediated DNA damage. Nat. Immunol. 20, 420–432 (2019).
Roulston, A. & Shore, G. C. New strategies to maximize therapeutic opportunities for NAMPT inhibitors in oncology. Mol. Cell Oncol. 3, e1052180 (2016).
Hong, S. M. et al. Increased nicotinamide adenine dinucleotide pool promotes colon cancer progression by suppressing reactive oxygen species level. Cancer Sci. 110, 629–638 (2019).
Piacente, F. et al. Nicotinic Acid Phosphoribosyltransferase Regulates Cancer Cell Metabolism, Susceptibility to NAMPT Inhibitors, and DNA Repair. Cancer Res 77, 3857–3869 (2017).
Ghanem, M. S. et al. Identification of NAPRT Inhibitors with Anti-Cancer Properties by In Silico Drug Discovery. Pharmaceuticals 15, (2022).
Belenky, P., Bogan, K. L. & Brenner, C. NAD+ metabolism in health and disease. Trends Biochem Sci. 32, 12–19 (2007).
Wu, X., Shats, I. & Li, X. Host-microbe interactions in NAD(+) metabolism. Trends Mol. Med, https://doi.org/10.1016/j.molmed.2025.07.001 (2025). Online ahead of print.
Shats, I. et al. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab. 31, 564–579 e567 (2020).
Chellappa, K. et al. NAD precursors cycle between host tissues and the gut microbiome. Cell Metab. 34, 1947–1959 e1945 (2022).
Su, Q. et al. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver. iScience 24, 103233 (2021).
Noah, T. K., Donahue, B. & Shroyer, N. F. Intestinal development and differentiation. Exp. Cell Res 317, 2702–2710 (2011).
Tung, K. L. et al. Integrated chromatin and transcriptomic profiling of patient-derived colon cancer organoids identifies personalized drug targets to overcome oxaliplatin resistance. Genes Dis. 8, 203–214 (2021).
Liu, C., Vyas, A., Kassab, M. A., Singh, A. K. & Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 (2017).
Luo, X. & Kraus, W. L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 (2012).
Yu, S. W. et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 (2002).
Zong, W. X., Ditsworth, D., Bauer, D. E., Wang, Z. Q. & Thompson, C. B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 18, 1272–1282 (2004).
Bai, P. et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 13, 461–468 (2011).
Rank, L. et al. Analyzing structure-function relationships of artificial and cancer-associated PARP1 variants by reconstituting TALEN-generated HeLa PARP1 knock-out cells. Nucleic Acids Res 44, 10386–10405 (2016).
Thomas, A. D. & Johnson, G. E. DNA Repair and Its Influence on Points of Departure for Alkylating Agent Genotoxicity. Thresholds of Genotoxic Carcinogens: From Mechanisms to Regulation, 67–82 (2016).
Yang, G. et al. Poly(ADP-ribosyl)ation mediates early phase histone eviction at DNA lesions. Nucleic Acids Res 48, 3001–3013 (2020).
Longarini, E. J. et al. Modular antibodies reveal DNA damage-induced mono-ADP-ribosylation as a second wave of PARP1 signaling. Mol. Cell 83, 1743–1760 e1711 (2023).
Zhang, L. Q. et al. Metabolic and molecular insights into an essential role of nicotinamide phosphoribosyltransferase. Cell Death Dis. 8, e2705 (2017).
Liu, L. et al. Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell Metab. 27, 1067–1080.e1065 (2018).
Nomura, M. et al. Niacin restriction with NAMPT-inhibition is synthetic lethal to neuroendocrine carcinoma. Nat. Commun. 14, 8095 (2023).
Song, W. S. et al. Nicotinic acid riboside maintains NAD(+) homeostasis and ameliorates aging-associated NAD(+) decline. Cell Metab. 37, 1616–1618 (2025).
Rosenberg, D. W., Giardina, C. & Tanaka, T. Mouse models for the study of colon carcinogenesis. Carcinogenesis 30, 183–196 (2009).
Hakem, R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 27, 589–605 (2008).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, 15 25 11-15 25 14 (2014).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Starr, A. E. et al. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn’s disease from UC. Gut 66, 1573–1583 (2017).
Moser, A. R., Pitot, H. C. & Dove, W. F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247, 322–324 (1990).
Su, L. K. et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670 (1992).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med 21, 1350–1356 (2015).
Gazzaniga, F., Stebbins, R., Chang, S. Z., McPeek, M. A. & Brenner, C. Microbial NAD Metabolism: Lessons from Comparative Genomics. Microbiol. Mol. Biol. Rev. 73, 529–541 (2009).
Hara, N. et al. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 282, 24574–24582 (2007).
Katsyuba, E. & Auwerx, J. Modulating N. A. D. + metabolism, from bench to bedside. EMBO J. 36, 2670–2683 (2017).
Wang, N. et al. NAPRT, but Not NAMPT, Provides Additional Support for NAD Synthesis in Esophageal Precancerous Lesions. Nutrients 14, 4916 (2022).
Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019).
Cohen, M. S. Interplay between compartmentalized NAD(+) synthesis and consumption: a focus on the PARP family. Genes Dev. 34, 254–262 (2020).
Ruszkiewicz, J. A., Burkle, A. & Mangerich, A. Fueling genome maintenance: On the versatile roles of NAD(+) in preserving DNA integrity. J. Biol. Chem. 298, 102037 (2022).
Tummala, K. S. et al. Inhibition of de novo NAD(+) synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 26, 826–839 (2014).
Gomes, A. L. et al. Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 30, 161–175 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Satija, R., Farrell, J., Gennert, D., Schier, A. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Yang, S. et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 21, 57 (2020).
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54, 985–995 (2022).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280 e6226 (2021).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013).
Liu, J. et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416 e411 (2018).
de Winter, J. C. F. Using the Student’s t-test with extremely small sample sizes. Pract. Assess. Res. Eval. 18, Article 10 (2013).
Acknowledgements
We thank NIEHS Comparative Medicine Branch for support to animal experiments; NIEHS Fluorescence Microscopy and Imaging Center for help with immunofluorescent analyses; NIEHS Epigenomics Core for bulk and single cell RNA sequencing; NIEHS Histology Core for tissue processing and staining; NIEHS Pathology Core laboratory for histopathological evaluation of DSS-colitis and AOM/DSS-induced CRC; NIEHS Flow Cytometry Center for assistance with FACS analysis; and the University of South Alabama Mass Spectrometry Core Facility for measurement of NUA. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH) to Xiaoling Li (Z01 ES102205). X. W. was supported in part by the NIH Office of Dietary Supplements (ODS) Scholars Award (TAS # 075-24-0846). Xiaojing Liu was supported by the National Institutes of Health Grant (GM150985). Joshua Hartsell (J. H.) was partially supported by a National Science Foundation STEM training grant (1643814). The contributions of the NIH authors were made as part of their official duties as NIH federal employees, are in compliance with agency policy requirements, and are considered Works of the United States Government. However, the findings and conclusions presented in this paper are those of the authors and do not necessarily reflect the views of the NIH or the U.S. Department of Health and Human Services.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
X. W. designed and performed experiments, analyzed data and wrote the manuscript. J. G. W performed LC-MS analysis of NAD metabolites in tissues. A. G. generated NAPRT KO mice. H. L., and Xiaojiang Xu analyzed bulk and single cell RNAseq data. J. H., J. S., and X. Liu performed LC-MS analysis of NAD metabolites in mouse plasma using LC-MS. R. M., C. D., M. J., and P. P. assisted with experiments. Y. F. generated the targeting plasmid for NAPRT inactivation in CRC119 cells. Xin Xu performed scRNAseq sequencing. Z. Z. and H. Y. assisted with organoid culture experiments. H. W. and A. K. J. performed metabolomic analysis. A. R. P. performed pathological evaluation of tumors in aged mice. M. E. M performed LC-MS analysis of NUA. J.-L. L. coordinated data analysis of scRNA-seq and RNA-seq and analyzed the expression of NAD metabolic enzymes in DC TCGA datasets and polyps datasets. I. S. designed and performed experiments, guided, designed, and coordinated the study, analyzed data and wrote the manuscript. X. Li guided, designed, and coordinated the study, analyzed data, and wrote the manuscript. All authors critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Nabil Djouder and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, X., Williams, J.G., Liang, H. et al. NAPRT-mediated deamidated NAD biosynthesis enhances colon tissue resiliency and suppresses tumorigenesis. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68998-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68998-w