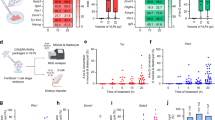
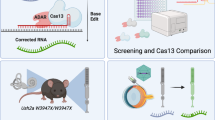
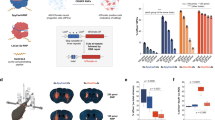
Abstract
CRISPR/Cas9-based gene-editing technologies offer promise for treating inherited retinal diseases (IRDs), however safe and efficient ocular delivery of precision editors remains challenging. To address this challenge, we report a class of Coomassie brilliant blue (CBB)-derived lipidoids that bind and deliver proteins. Subretinal injection of Cre complexed with these lipidoids into mT/mG mice leads to robust recombination in the retinal pigment epithelium and photoreceptors. We employ the CBB-lipidoid platform to deliver adenine base editor (ABE) ribonucleoproteins (RNP). Incorporating CBB lipidoids into liposomes improves delivery efficiency. CBB11 stands out for facilitating precise in vivo ABE-mediated gene editing. Delivery of liposome-CBB11-RNP complexes results in a 120-fold increase in base editing compared to RNP alone and restores the scotopic ERG b-wave response in the rd12 mouse model. These results demonstrate the potential of CBB-augmented, liposome-RNP systems for therapeutic gene editing in the eye, paving the way for single-dose precision medicines to treat IRDs.
Similar content being viewed by others
Data availability
The high-throughput sequencing data generated in this study were deposited in the National Center for Biotechnology Information Sequence Read Archive database under accession codes: PRJNA1240374 and PRJNA1358750. The data underlying the manuscript were deposited in Dryad: doi.org/10.5061/dryad.d51c5b0hd54 [https://datadryad.org/dataset/doi:10.5061/dryad.d51c5b0hd]. Source data are provided in this paper.
References
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Wang, J. H., Gessler, D. J., Zhan, W., Gallagher, T. L. & Gao, G. Adeno-associated virus as a delivery vector for gene therapy of human diseases. Signal Transduct. Target. Ther. 9, 78 (2024).
Jang, H. et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 6, 181–194 (2022).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010).
Fehrman, R. L., Chern, K. J., Stoltz, K. P. & Lipinski, D. M. The vectors went in two-by-two: Transduction efficiency and tolerability of dual and triple rAAV vector delivery following intravitreal injection for genome-editing applications. Exp. Eye Res. 251, 110223 (2025).
Padilla, M. S. et al. Branched endosomal disruptor (BEND) lipids mediate delivery of mRNA and CRISPR-Cas9 ribonucleoprotein complex for hepatic gene editing and T cell engineering. Nat. Commun. 16, 996 (2025).
Lian, X. et al. Bone-marrow-homing lipid nanoparticles for genome editing in diseased and malignant haematopoietic stem cells. Nat. Nanotechnol. 19, 1409–1417 (2024).
Dong, S. et al. Lipid nanoparticles-mediated mRNA delivery to the eye affected by ionizable cationic lipid. Mol. Pharm. 22, 3297–3307 (2025).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Wei, T. et al. Lung SORT LNPs enable precise homology-directed repair mediated CRISPR/Cas genome correction in cystic fibrosis models. Nat. Commun. 14, 7322 (2023).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Holubowicz, R. et al. Safer and efficient base editing and prime editing via ribonucleoproteins delivered through optimized lipid-nanoparticle formulations. Nat. Biomed. Eng. 9, 57–78 (2025).
Chen, K. et al. Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR-Cas9 ribonucleoprotein. Nat. Biotechnol. 43, 1445–1457 (2025).
Tsuchida, C. A., Wasko, K. M., Hamilton, J. R. & Doudna, J. A. Targeted nonviral delivery of genome editors in vivo. Proc. Natl. Acad. Sci. USA 121, e2307796121 (2024).
Tao, Y. et al. Treatment of monogenic and digenic dominant genetic hearing loss by CRISPR-Cas9 ribonucleoprotein delivery in vivo. Nat. Commun. 14, 4928 (2023).
Tan, Z. et al. Block Polymer Micelles Enable CRISPR/Cas9 Ribonucleoprotein Delivery: Physicochemical Properties Affect Packaging Mechanisms and Gene Editing Efficiency. Macromolecules 52, 8197–8206 (2019).
Metzger, J. M. et al. Efficient in vivo neuronal genome editing in the mouse brain using nanocapsules containing CRISPR-Cas9 ribonucleoproteins. Biomaterials 293, 121959 (2023).
Liu, C. et al. A boronic acid-rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 5, eaaw8922 (2019).
Wei, T., Cheng, Q., Min, Y. L., Olson, E. N. & Siegwart, D. J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020).
Lee, B. et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2, 497–507 (2018).
Felgner, P. L. et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 84, 7413–7417 (1987).
Cullis, P. R. & Felgner, P. L. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat. Rev. Drug Discov. 23, 709–722 (2024).
Cho, E. Y. et al. Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes. J. Nanobiotechnol. 17, 19 (2019).
Riley, R. S. et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv. 7, https://doi.org/10.1126/sciadv.aba1028 (2021).
Han, X. et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 12, 7233 (2021).
Zhang, Y., Sun, C., Wang, C., Jankovic, K. E. & Dong, Y. Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021).
Georgiou, C. D., Grintzalis, K., Zervoudakis, G. & Papapostolou, I. Mechanism of Coomassie brilliant blue G-250 binding to proteins: a hydrophobic assay for nanogram quantities of proteins. Anal. Bioanal. Chem. 391, 391–403 (2008).
Tai, W., Zhao, P. & Gao, X. Cytosolic delivery of proteins by cholesterol tagging. Sci. Adv. 6, eabb0310 (2020).
Hołubowicz, R. et al. Precision base and prime editing in the eye using ribonucleoprotein nanoparticles. Nat. Biomed. Eng. 9, 57–78 (2024).
Du, S. W. et al. TIGER: A tdTomato in vivo genome-editing reporter mouse for investigating precision-editor delivery approaches. Proc. Natl. Acad. Sci. USA 122, e2506257122 (2025).
Chen, L. et al. Engineering a precise adenine base editor with minimal bystander editing. Nat. Chem. Biol. 19, 101–110 (2023).
Li, J. et al. Comprehensive single-cell atlas of the mouse retina. iScience 27, 109916 (2024).
Suh, S. et al. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 5, 169–178 (2021).
Choi, E. H. et al. In vivo base editing rescues cone photoreceptors in a mouse model of early-onset inherited retinal degeneration. Nat. Commun. 13, 1830 (2022).
Banskota, S. et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 185, 250–265 (2022).
Arantes, P. R. et al. Dimerization of the deaminase domain and locking interactions with Cas9 boost base editing efficiency in ABE8e. Nucleic Acids Res. 52, 13931–13944 (2024).
Lapinaite, A. et al. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science 369, 566–571 (2020).
Keane, P. A. et al. Objective measurement of vitreous inflammation using optical coherence tomography. Ophthalmology 121, 1706–1714 (2014).
Fliesler, S. J. & Anderson, R. E. Chemistry and metabolism of lipids in the vertebrate retina. Prog. Lipid Res. 22, 79–131 (1983).
Hamilton, A. G. et al. High-Throughput In Vivo Screening Identifies Differential Influences on mRNA Lipid Nanoparticle Immune Cell Delivery by Administration Route. ACS Nano 18, 16151–16165 (2024).
Pulman, J. et al. Direct delivery of Cas9 or base editor protein and guide RNA complex enables genome editing in the retina. Mol. Ther. Nucleic Acids 35, 102349 (2024).
An, M. et al. Engineered virus-like particles for transient delivery of prime editor ribonucleoprotein complexes in vivo. Nat. Biotechnol. 42, 1526–1537 (2024).
Du, S. W. et al. In vivo photoreceptor base editing ameliorates rhodopsin-E150K autosomal-recessive retinitis pigmentosa in mice. Proc. Natl. Acad. Sci. USA 121, e2416827121 (2024).
Geilenkeuser, J. et al. Engineered nucleocytosolic vehicles for loading of programmable editors. Cell188, 2637–2655 (2025).
Mazzoni, F., Tombo, T. & Finnemann, S. C. No difference between age-matched male and female C57BL/6J mice in photopic and scotopic electroretinogram a- and b-wave amplitudes or in peak diurnal outer segment phagocytosis by the retinal pigment epithelium. Adv. Exp. Med. Biol. 1185, 507–511 (2019).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Palczewska, G., Kern, T. S. & Palczewski, K. Noninvasive two-photon microscopy imaging of mouse retina and retinal pigment epithelium. Methods Mol. Biol. 1834, 333–343 (2019).
Palczewska, G. et al. Noninvasive two-photon microscopy imaging of mouse retina and retinal pigment epithelium through the pupil of the eye. Nat. Med. 20, 785–789 (2014).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Zhang, J. et al. A combinatorial synthetic strategy for developing genome-editing protein-delivery agents targeting mouse retina, Dataset in Dryad, https://doi.org/10.5061/dryad.d51c5b0hd (2025).
Acknowledgements
We thank Marco Bassetto, Elliot H. Choi, Huajun B. Yan, and Alexander L. Yan for technical assistance. We acknowledge Dorota Skowronska-Krawczyk for access to a qPCR thermocycler, fluorescent microscope, and gel imager. We thank Suvrajit Sengupta at the UCI Department of Chemistry for technical support with NMR. We thank Jennifer Atwood and Michael Hou for technical assistance with flow cytometry. We thank Yekaterina Kadyshevskaya (University of Southern California) for the preparation of Fig. 1. We thank our colleagues at the UCI Center for Translational Vision Research and the Gavin Herbert Eye Institute for their comments regarding this manuscript. This work was supported in part by grants from the National Institutes of Health, including R01EY009339 (K.P.), 1R01EY034501 (K.P.), NSF-CHE-1904530 (G.P.T.), NIAID 75N93022C00054 (P.L.F.), DTRA grant N66001-21-C-4013 (P.L.F.), UG3AI150551 (D.R.L.), U01AI142756 (D.R.L.), R35GM118062 (D.R.L.), RM1HG009490 (D.R.L.), T32GM008620 (S.W.D.), F30EY033642 (S.W.D.); Foundation Fighting Blindness (award number TA-GT-0423-0847-UCI-TRAP) (K.P.); the Howard Hughes Medical Institute (HHMI) (D.R.L.); and Knights Templar Eye Foundation Career-Starter Research Grant (R.H.). R.H. is a Beckman Scholar in Retinal Research. The authors acknowledge support to the Department of Ophthalmology, Gavin Herbert Eye Institute at the University of California, Irvine, from an unrestricted Research to Prevent Blindness award, from NIH core grant P30EY034070, and from a University of California, Irvine School of Medicine Dean’s Office grant. The authors acknowledge support for the Chao Family Comprehensive Cancer Center’s Institute for Immunology Flow Cytometry Facility shared resource by the National Cancer Institute of the National Institutes of Health under award number P30CA062203. This article is subject to the Open Access to Publications policy of the Howard Hughes Medical Institute (HHMI). HHMI-supported authors have previously granted a nonexclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles. Pursuant to those licenses, the author-accepted version of this manuscript can be made freely available under a CC BY 4.0 license immediately upon publication.
Author information
Authors and Affiliations
Contributions
Conceptualization was performed by J.Z., G.P.T, P.L.F., and K.P. Experimental investigations were performed by J.Z., S.W.D., J.H.F., R.S., R.H., G. P., G.P.T., C.R.M., Y.H., E.R., Z.D., X.M., M.H.S., L. X., M.H., P.Z.C., and B.L. Data analysis was performed by J.Z., R.H., S.W.D., J.H.F., R.S., C.R.M., E.R., G.P., and Y.H. Figures were prepared by J.Z., R.H., Y.H., G.P., and G.P.T. Manuscript was written by J.Z., R.H., Y.H., G.P.T., and K.P. Project was supervised by D.R.L., G.P.T., and K.P. Funds were acquired by R.H., S.W.D., D.R.L., G.P.T, P.L.F., and K.P. All authors contributed to the research and writing, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.P. is a consultant for Polgenix Inc. and AbbVie Inc., and serves on the Scientific Advisory Board of Hyperion Eye Ltd. K.P. and G.P.T. are equity holders in Eyesomer Therapeutics. D.R.L. is a consultant and/or equity owner for Prime Medicine, Beam Therapeutics, Pairwise Plants, and nChroma Bio, companies that use or deliver genome-editing or epigenome-engineering agents. All other authors have declared that no competing interests exist.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, J., Hołubowicz, R., Smidak, R. et al. A combinatorial synthetic strategy for developing genome-editing protein-delivery agents targeting mouse retina. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69077-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69077-w