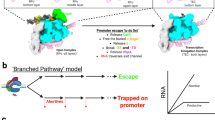
Abstract
In bacteria, RNA polymerase (RNAP) often pauses during the early stages of transcription initiation. The structural basis for these transient pauses remains unclear. Here, we present cryo-electron microscopy (cryo-EM) structures of the paused initiation complex (PIC) and initiation complex (IC) of Mycobacterium tuberculosis (Mtb), which include the RNAP core enzyme, the ECF σ factor σE, transcription factor CarD, promoter DNA, and nascent RNA. Our structures with pre-melted scaffolds reveal an intermediate at the 6–7 nt stage compatible with a paused-like intermediate, associated with steric hindrance between the emerging RNA and the σ3.2 region. This clash triggers a swivel of the RNAP structural module and scrunching of the transcription bubble. We also observe positional rearrangement of the σ4 domain, suggesting a poised pre-escape state. In addition, complementary reconstructions with fully matched DNA scaffolds (N-IC and N-PIC) support the physiological relevance of the captured intermediates. Together, our results support the existence of a mechanistic checkpoint during transcription initiation and suggest an RNA-induced model how RNAP conformational dynamics regulate early transcription.
Similar content being viewed by others
Data availability
The Cryo-EM density maps and structures have been deposited into the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) with the accession numbers 9M98 and EMD-63729 for Mtb IC6, 9M9D and EMD-63730 for Mtb IC7, 9M9E and EMD-63731 for Mtb PIC7, EMD-66362 for N-PIC7, EMD-66363 for N-IC6, and EMD-66364 for N-IC7. The PDB entries 8E8M, 8E82, 5UH5, 5ZX2, 6DVD, 6EDT, 6JBQ and 6JCY have been used for structure comparison in this study. Source data are provided with this paper.
References
Goovaerts, Q. et al. Structures illustrate step-by-step mitochondrial transcription initiation. Nature 622, 872–879 (2023).
Chen, X. Z. et al. Structural visualization of transcription initiation in action. Science 382, 1–13 (2023).
He, Y., Fang, J., Taatjes, D. J. & Nogales, E. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013).
Feklistov, A. & Darst, S. A. Structural basis for promoter-10 element recognition by the bacterial rna polymerase σ subunit. Cell 147, 1257–1269 (2011).
Feklístov, A., Sharon, B. D., Darst, S. A. & Gross, C. A. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 (2014).
Boyaci, H., Chen, J., Jansen, R., Darst, S. A. & Campbell, E. A. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature 565, 382–385 (2019).
Ruff, E., Record, M. & Artsimovitch, I. Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015).
Chen, J., Boyaci, H. & Campbell, E. A. Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 19, 95–109 (2020).
Abdelkareem, M. et al. Structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol. Cell 75, 298–309 (2019).
Cheung, A. C. M. & Cramer, P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 (2011).
Duchi, D. et al. RNA polymerase pausing during initial transcription. Mol. Cell 63, 939–950 (2016).
Dulin, D. et al. Pausing controls branching between productive and non-productive pathways during initial transcription in bacteria. Nat. Commun. 9, 1–11 (2018).
Lerner, E. et al. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 113, E6562–E6571 (2016).
Lonetto, M. A., Brown, K. L., Rudd, K. E. & Buttner, M. J. Analysis of the streptomyces-coelicolor SigE gene reveals the existence of a subfamily of eubacterial RNA-polymerase sigma-factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA. 91, 7573–7577 (1994).
Missiakas, D. & Raina, S. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28, 1059–1066 (1998).
Helmann, J. D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46, 47–110 (2002).
Helmann, J. D. extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr. Opin. Microbiol. 30, 122–132 (2016).
Casas-Pastor, D. et al. Expansion and re-classification of the extracytoplasmic function (ECF) σ factor family. Nucleic Acids Res 49, 986–1005 (2021).
Sineva, E., Savkina, M. & Ades, S. E. Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 36, 128–137 (2017).
Murakami, K. S., Masuda, S. & Darst, S. A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296, 1280–1284 (2002).
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002).
Mekler, V. et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108, 599–614 (2002).
Basu, R. S. et al. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 289, 24549–24559 (2014).
Li, L. T., Molodtsov, V., Lin, W., Ebright, R. H. & Zhang, Y. RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription. Proc. Natl. Acad. Sci. USA. 117, 5801–5809 (2020).
Pukhrambam, C. et al. Structural and mechanistic basis of σ-dependent transcriptional pausing. Proc. Natl. Acad. Sci. USA 119, e2201301119 (2022).
Bae, B., Feklistov, A., Lass-Napiorkowska, A., Landick, R. & Darst, S. A. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4, e08504 (2015).
Winkelman, J. T., Nickels, B. E. & Ebright, R. H. “The Transition from Transcription Initiation to Transcription Elongation: Start-site Selection, Initial Transcription, and Promoter Escape” In RNA Polymerases as a Molecular Motor, Landick, R. W., Strick, T., Eds. (RSC Publishing, 2019).
Kapanidis, A. N. et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006).
Revyakin, A., Liu, C. Y., Ebright, R. H. & Strick, T. R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006).
Roberts, J. W. & Biochemistry, - RNA polymerase, a scrunching machine. Science 314, 1097–1098 (2006).
Kramm, K., Endesfelder, U. & Grohmann, D. A single-molecule view of archaeal transcription. J. Mol. Biol. 431, 4116–4131 (2019).
Winkelman, J. T. et al. Multiplexed protein-DNA cross-linking: scrunching in transcription start site selection. Science 351, 1090–1093 (2016).
Fang, C. L. et al. Structures and mechanism of transcription initiation by bacterial ECF factors. Nucleic Acids Res. 47, 7094–7104 (2019).
Mascher, T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 16, 148–155 (2013).
Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L. & Manganelli, R. The σ factors of mycobacterium tuberculosis. FEMS Microbiol. Rev. 30, 926–941 (2006).
Manganelli, R., Voskuil, M. I., Schoolnik, G. K. & Smith, I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437 (2001).
Donà, V. et al. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor sigmaE in Mycobacterium tuberculosis. J. Bacteriol. 190, 5963–5971 (2008).
Hausnerová, V. V. et al. RIP-seq reveals RNAs that interact with RNA polymerase and primary sigma factors in bacteria. Nucleic Acids Res. 52, 4604–4626 (2024).
Manganelli, R. et al. SigE: a master regulator of Mycobacterium tuberculosis. Front. Microbiol. 14, 1–8 (2023).
Li, L. T., Fang, C. L., Zhuang, N. N., Wang, T. T. & Zhang, Y. Structural basis for transcription initiation by bacterial ECF σ factors. Nat. Commun. 10, 1–14 (2019).
Lin, W. et al. Structural basis of ECF-σ-factor-dependent transcription initiation. Nat. Commun. 10, 1–14 (2019).
Bae, B. et al. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. eLife 4, e08505 (2015).
Stallings, C. L. et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159 (2009).
Zhu, D. X. & Stallings, C. L. Transcription regulation by CarD in mycobacteria is guided by basal promoter kinetics. J. Biol. Chem. 299, 104724 (2023).
Abellón-Ruiz, J. et al. The CarD/CarG regulatory complex is required for the action of several members of the large set of extracytoplasmic function σ factors. Environ. Microbiol. 16, 2475–2490 (2014).
Delbeau, M. et al. Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis. Mol. Cell 83, 1474–1488 (2023).
Guo, X. Y. et al. Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827 (2018).
Vishwakarma, R. K., Qayyum, M. Z., Babitzke, P. & Murakami, K. S. Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase. Proc. Natl. Acad. Sci. USA. 120, e2218516120 (2023).
Cordes, T. et al. Sensing DNA opening in transcription using quenchable Forster resonance energy transfer. Biochemistry 49, 9171–9180 (2010).
Rammohan, J. et al. Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD. Nucleic Acids Res 44, 7304–7313 (2016).
Chauvier, A. et al. Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand. Nat. Struct. Mol. Biol. 30, 902–913 (2023).
Dey, S. et al. Structural insights into RNA-mediated transcription regulation in bacteria. Mol. Cell 82, 3885–3900 (2022).
Imashimizu, M. et al. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J. Mol. Biol. 425, 697–712 (2013).
Robb, N. C. et al. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection. J. Mol. Biol. 425, 875–885 (2013).
Chakraborty, A. et al. Opening and closing of the bacterial RNA polymerase clamp. Science 337, 591–595 (2012).
Alley, S. C. et al. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38, 7696–7709 (1999).
Mazumder, A., Ebright, R. H. & Kapanidis, A. N. Transcription initiation at a consensus bacterial promoter proceeds via a ‘bind-unwind-load-and-lock’ mechanism. eLife 10, e70090 (2021).
Wang, D., Bushnell, D. A., Westover, K. D., Kaplan, C. D. & Kornberg, R. D. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006).
Kang, J. Y. et al. An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options. Proc. Natl. Acad. Sci. USA. 120, e2215945 (2023).
Zhu, C. J. et al. Transcription factors modulate RNA polymerase conformational equilibrium. Nat. Commun. 13, 1–12 (2022).
Kang, J. Y. et al. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815 (2018).
Bao, Y. & Landick, R. Obligate movements of an active site-linked surface domain control RNA polymerase elongation and pausing via a Phe pocket anchor. Proc. Natl. Acad. Sci. USA. 118, e2101805118 (2021).
Gao, F. et al. Structural basis of sigma54 displacement and promoter escape in bacterial transcription. Proc. Natl Acad. Sci. USA. 121, e2309670120 (2024).
Wang, A. et al. The displacement of the sigma70 finger in initial transcription is highly heterogeneous and promoter-dependent. Nucleic Acids Res. 53, 857 (2025).
Hazra, J. P. et al. Unraveling single-molecule reactions via multiplexed in-situ DNA sequencing. Preprint at https://doi.org/10.1101/2025.11.27.690701, (2025).
Winkelman, J. T. et al. XACT-Seq comprehensively defines the promoter-position and promoter-sequence determinants for initial-transcription pausing. Mol. Cell 79, e798 (2020).
Wang, C. Y. et al. Structural basis of transcription-translation coupling. Science 369, 1359–1365 (2020).
Webster, M. W. et al. Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359 (2020).
O’Reilly, F. J. et al. In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557 (2020).
Said, N. et al. Structural basis for λN-dependent processive transcription antitermination. Nat. Microbiol. 2, 17062 (2017).
Bar-Nahum, G. & Nudler, E. Isolation and characterization of sigma(70)-retaining transcription elongation complexes from Escherichia coli. Cell 106, 443–451 (2001).
Harden, T. T. et al. Bacterial RNA polymerase can retain sigma70 throughout transcription. Proc. Natl. Acad. Sci. USA. 113, 602–607 (2016).
Kapanidis, A. N. et al. Retention of transcription initiation factor sigma70 in transcription elongation: single-molecule analysis. Mol. Cell 20, 347–356 (2005).
Ju, X. W. et al. Incomplete transcripts dominate the Mycobacterium tuberculosis transcriptome. Nature 627, 424–430 (2024).
Wang, Q., Ren, Y., Jin, Q., Chen, X. & Xu, Y. Structural insights into human Pol III transcription initiation in action. Nature 643, 1127–1134 (2025).
Dolgosheina, E. V. et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 9, 2412–2420 (2014).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Acknowledgements
We thank the staff members at the National Facility for Protein Science in Shanghai Zhangjiang Lab and the cryo-EM centers of CAS Center for Excellence in Molecular Plant Sciences for their technical assistance on cryo-EM data collection. This study was funded by grants from the National Natural Science Foundation of China (32500154 to L.Z.; 32301018 and 32571394 to K.X.), Shanghai Sailing Program (23YF1450100 to L.Z.), Shanghai Health Commission Clinical Research Special Youth Project (20234Y0116 to L.Z.), Tongji University Independent Original Basic Research Project (22120240275 to L.Z.), the National Key Research and Development Program of China (2021YFA1302200 to K.X.), and the Thousand Talents Plan-Youth to K.X., Fundamental Research Funds for the Central Universities (Tongji University) to K.X.
Author information
Authors and Affiliations
Contributions
L.Z. conceived of the project and designed the experiments. L.Z. performed the sample purification and complex assembly. L.Z. collected the cryo-EM data. L.Z. performed Fluorescence-detecting in vitro transcription assays. L.Z. and K.X. analyzed the data. L.Z. and K.X. wrote the manuscript. All authors discussed the experiments, read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Konstantin Brodolin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zheng, L., Xu, K. Structural basis of pausing during transcription initiation in mycobacterium tuberculosis. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69104-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69104-w