Abstract
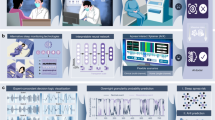
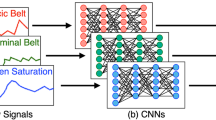
Diagnosing sleep disordered breathing requires manual annotation of events from sleep studies, such as nocturnal polysomnography, a process that is time-intensive, costly, and prone to inter-rater variability. Automatic approaches exist but lack generalizability due to signal variability across centers. We develop an automatic apneic breathing event detector to localize and classify obstructive apneas, central apneas, hypopneas, and isolated respiratory events without arousals or desaturations. The model is trained on 5456 polysomnographies and tested on 1099 polysomnographies from six cohorts uses an end-to-end deep learning architecture. The model’s predictions show a strong correlation with expert annotations for apnea-hypopnea index (r² = 0.84) and achieve an F1 score of 0.78 across apnea event types, with specific F1 scores of 0.71, 0.51, and 0.65 for obstructive apnea, central apnea, and hypopnea events, respectively. In two independent, multi-scored datasets, The model performs comparably or better than individual expert raters. The model’s probabilistic output, termed “apnotyping,” provides insights into sleep disordered breathing etiology, with event probabilities correlating more strongly with key sleep apnea traits—such as loop gain and pharyngeal muscle compensation—than traditional apnea indexes. This probabilistic approach may enhance diagnostic accuracy and support personalized treatment strategies, leading to improved patient outcomes.
Similar content being viewed by others
Data availability
The polysomnography data from the DREEM dataset are publicly available via Zenodo (https://zenodo.org/records/15900394). The remaining polysomnography data in this study are available under restricted access due to ethical and legal constraints. Data from the Multi-Ethnic Study of Atherosclerosis (MESA), the Osteoporotic Fractures in Men Study (MrOS), the Cleveland Family Study (CFS), Wisconsin Sleep Cohort (WSC), and ALLIANCE can be obtained through the National Sleep Research Resource (NSRR; https://sleepdata.org) following data use agreement approval. The data will be made available within a month through the NSRR and is then available for the expected duration of the study. Source data are provided with this paper.
Code availability
All custom source code, along with software packages and versions used for this project, is available at https://github.com/RuudeResearch/ABED.
References
American Academy of Sleep Medicine. International classification of sleep disorders: Diagnostic and coding manual, third edition. American Academy of Sleep Medicine https://learn.aasm.org/Public/Catalog/Details.aspx?id=%2FgqQVDMQIT%2FEDy86PWgqgQ%3D%3D&returnurl=%2FUsers%2FUserOnlineCourse.aspx%3FLearningActivityID%3D%252fgqQVDMQIT%252fEDy86PWgqgQ%253d%253d (2014).
L‚vy, P. et al. Obstructive sleep apnoea syndrome. Nat. Rev. Dis. Prim. 1, 15015 (2015).
Berry, R. B. et al. AASM | Scoring Manual Version 2.2 The AASM Manual for the Scoring of Sleep and Associated Events Rules, Terminology And Technical Specifications Version 2.2. www.aasmnet.org (2015).
Stanchina, M. et al. Clinical use of loop gain measures to determine continuous positive airway pressure efficacy in patients with complex sleep apnea. a pilot study. Ann. Am. Thorac. Soc. 12, 1351–1357 (2015).
Hirotsu, C. et al. Effect of three hypopnea scoring criteria on OSA prevalence and associated comorbidities in the general population. J. Clin. Sleep. Med 15, 183–194 (2019).
Berry, R. B. et al. A transition to the American Academy of Sleep Medicine-recommended hypopnea definition in adults: initiatives of the Hypopnea Scoring Rule Task Force. J. Clin. Sleep. Med 18, 1419–1425 (2022).
Benjafield, A. V. et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir. Med. 7, 687 (2019).
Kuna, S. T. et al. Agreement in computer-assisted manual scoring of polysomnograms across sleep centers. Sleep 36, 583–589 (2013).
Blaha, M. J. & DeFilippis, A. P. Multi-Ethnic Study of Atherosclerosis (MESA): JACC focus seminar 5/8. J. Am. Coll. Cardiol. 77, 3195–3216 (2021).
Khan, A. et al. Obstructive sleep apnea during rapid eye movement sleep, daytime sleepiness, and quality of life in older men with osteoporotic fractures in men (MrOS) sleep study. J. Clin. Sleep Med. 9, 191–198 (2013).
Sankari, A. et al. Characteristics of non-apneic respiratory events-sankari et al. Characteristics and consequences of non-apneic respiratory events during sleep. Sleep 40, zsw024 (2017).
Malhotra, A. et al. Metrics of sleep apnea severity: beyond the apnea-hypopnea index. Sleep 44, zsab030 (2021).
Pinilla, L. et al. Hypoxic burden to guide CPAP treatment allocation in patients with obstructive sleep apnoea: a post hoc study of the ISAACC trial. Eur. Respir. J. 62, 2300828 (2023).
Labarca, G. et al. Sleep apnea physiological burdens and cardiovascular morbidity and mortality. Am. J. Respir. Crit. Care Med 208, 802–813 (2023).
Esmaeili, N. et al. Hypoxic burden based on automatically identified desaturations is associated with adverse health outcomes. Ann. Am. Thorac. Soc. 20, 1633–1641 (2023).
Koch, H. et al. Breathing disturbances without hypoxia are associated with objective sleepiness in sleep apnea. Sleep 40, zsx152 (2017).
Mann, D. L. et al. Flow limitation is associated with excessive daytime sleepiness in individuals without moderate or severe obstructive sleep apnea. Ann. Am. Thorac. Soc. 21, 1186–1193 (2024).
Azarbarzin, A. et al. Relevance of cortical arousals for risk stratification in sleep apnea: a 3-cohort analysis. J. Clin. Sleep. Med 19, 1475–1484 (2023).
Kjar, M. R. et al. Polysomnographic plethysmography excursions are reduced in obese elderly men. Annu Int Conf. IEEE Eng. Med Biol. Soc. 2021, 2396–2399 (2021).
Malhotra, A., Mesarwi, O., Pepin, J. L. & Owens, R. L. Endotypes and phenotypes in obstructive sleep apnea. Curr. Opin. Pulm. Med 26, 609 (2020).
Eckert, D. J., White, D. P., Jordan, A. S., Malhotra, A. & Wellman, A. Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets. Am. J. Respir. Crit. Care Med 188, 996–1004 (2013).
Azarbarzin, A. Estimation of pharyngeal collapsibility during sleep by peak inspiratory airflow. Sleep 40, zsw005 (2017).
Terrill, P. I. et al. Quantifying the ventilatory control contribution to sleep apnoea using polysomnography. Eur. Respir. J. 45, 408–418 (2015).
Anwar, A. I. et al. Novel physiologic predictors of positive airway pressure effectiveness (NICEPAP) study: rationale, design and methods. Sleep. Breath. 28, 2005–2015 (2024).
Dutta, R. et al. A novel model to estimate key obstructive sleep apnea endotypes from standard polysomnography and clinical data and their contribution to obstructive sleep apnea severity. Ann. Am. Thorac. Soc. 18, 656–667 (2021).
Alex, R. M. et al. Within-night repeatability and long-term consistency of sleep apnea endotypes: the Multi-Ethnic Study of Atherosclerosis and Osteoporotic Fractures in Men Study. Sleep 45, zsac129 (2022).
Chu, Y. & Zinchuk, A. The present and future of the clinical use of physiological traits for the treatment of patients with OSA: a narrative review. J. Clin. Med. 13, 1636 (2024).
Aishah, A. et al. Stepwise add-on and endotype-informed targeted combination therapy to treat obstructive sleep apnea: a proof-of-concept study. Ann. Am. Thorac. Soc. 20, 1316–1325 (2023).
Rosenberg, R. S. & Van Hout, S. The American Academy of Sleep Medicine inter-scorer reliability program: respiratory events. J. Clin. Sleep. Med. 10, 447–454 (2014).
Urtnasan, E., Park, J. U. & Lee, K. J. Automatic detection of sleep-disordered breathing events using recurrent neural networks from an electrocardiogram signal. Neural Comput. Appl. 32, 4733–4742 (2020).
Tripathy, R. K., Gajbhiye, P. & Acharya, U. R. Automated sleep apnea detection from cardio-pulmonary signal using bivariate fast and adaptive EMD coupled with cross-time-frequency analysis. Comput. Biol. Med. 120, 103769 (2020).
Van Steenkiste, T., Groenendaal, W., Deschrijver, D. I. & Dhaene, T. Automated sleep apnea detection in raw respiratory signals using long short-term memory neural networks. IEEE J. Biomed. Health Inf. 23, 2354–2364 (2019).
Zhao, X. et al. Classification of sleep apnea based on EEG sub-band signal characteristics. Sci. Rep. 2021 11, 1–11 (2021).
Mostafa, S. S., Mendonca, F., Ravelo-Garcia, A. G., Julia-Serda, G. & Morgado-Dias, F. Multi-objective hyperparameter optimization of a convolutional neural network for obstructive sleep apnea detection. IEEE Access 8, 129586–129599 (2020).
Yu, H. et al. A sleep apnea-hypopnea syndrome automatic detection and subtype classification method based on LSTM-CNN. Biomed. Signal Process Control 71, 103240 (2022).
Biswal, S. et al. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 25, 1643–1650 (2018).
Haidar, R., McCloskey, S., Koprinska, I. & Jeffries, B. Convolutional neural networks on multiple respiratory channels to detect hypopnea and obstructive apnea events. Proceedings of the International Joint Conference on Neural Networks. 2018-July, (2018).
Zahid, A. N., Jennum, P., Mignot, E. & Sorensen, H. B. D. MSED: a multi-modal sleep event detection model for clinical sleep analysis. IEEE Trans. Biomed. Eng. 70, 2508–2518 (2023).
Yeo, M. et al. Respiratory event detection during sleep using electrocardiogram and respiratory-related signals: using polysomnogram and patch-type wearable device data. IEEE J. Biomed. Health Inf. 26, 550–560 (2022).
Lakhan, P., Ditthapron, A., Banluesombatkul, N. & Wilaiprasitporn, T. Deep neural networks with weighted averaged overnight airflow features for sleep apnea-hypopnea severity classification. IEEE Region 10 Annual International Conference, Proceedings/TENCON 2018-October, 441–445 (2018).
Nassi, T. E. et al. Automated scoring of respiratory events in sleep with a single effort belt and deep neural networks. IEEE Trans. Biomed. Eng. 69, 2094 (2022).
Olesen, A. N., Jorgen Jennum, P., Mignot, E. & Sorensen, H. B. D. Automatic sleep stage classification with deep residual networks in a mixed-cohort setting. Sleep 44, zsaa161 (2021).
Cohen, O. et al. Achieving better understanding of obstructive sleep apnea treatment effects on cardiovascular disease outcomes through machine learning approaches: a narrative review. J. Clin. Med. 13, 1415 (2024).
Dean, D. A. et al. Scaling up scientific discovery in sleep medicine: the national sleep research resource. Sleep 39, 1151–1164 (2016).
Brink-Kjaer, A. et al. Automatic detection of cortical arousals in sleep and their contribution to daytime sleepiness. Clin. Neurophysiol. 131, 1187–1203 (2020).
Moon, K. R. et al. PHATE: A dimensionality reduction method for visualizing trajectory structures in high-dimensional biological data. bioRxiv https://doi.org/10.1101/120378 (2017).
Guillot, A., Sauvet, F., During, E. H. & Thorey, V. Dreem open datasets: multi-scored sleep datasets to compare human and automated sleep staging. IEEE Trans. Neural Syst. Rehab. Eng. 28, 1955–1965 (2020).
Sands, S. A. et al. Pathophysiology underlying demographic and obesity determinants of sleep apnea severity. Ann. Am. Thorac. Soc. 20, 440–449 (2023).
Faria, A., Allen, A. H., Fox, N., Ayas, N. & Laher, I. The public health burden of obstructive sleep apnea. Sleep. Sci. 14, 257 (2021).
Levy, J., Álvarez, D., Del Campo, F. & Behar, J. A. Deep learning for obstructive sleep apnea diagnosis based on single channel oximetry. Nat. Commun. 14, 1–12 (2023).
Eckert, D., Dutta, R. & Levendowski, D. 0602 Application of a physiology-based OSA endotype model to predict oral appliance outcomes using home. Sleep. Study Data. Sleep. 47, A257–A257 (2024).
Dutta, R., Tong, B. K. & Eckert, D. J. Development of a physiological-based model that uses standard polysomnography and clinical data to predict oral appliance treatment outcomes in obstructive sleep apnea. J. Clin. Sleep. Med. 18, 861–870 (2022).
Hang, L.-W. et al. Sex-specific age-related worsening of pathological endotypic traits in patients with obstructive sleep apnea. Sleep https://doi.org/10.1093/SLEEP/ZSAE185 (2024).
Sands, S. A. Identifying obstructive sleep apnoea patients responsive to supplemental oxygen therapy. Eur. Respir. J. 52, 1800674 (2018).
Joosten, S. A. Loop gain predicts the response to upper airway surgery in patients with obstructive sleep apnea. Sleep https://doi.org/10.1093/sleep/zsx094 (2017).
Bamagoos, A. A. et al. Polysomnographic endotyping to select patients with obstructive sleep apnea for oral appliances. Ann. Am. Thorac. Soc. 16, 1422–1431 (2019).
Op de Beeck, S. et al. Endotypic mechanisms of successful hypoglossal nerve stimulation for obstructive sleep apnea. Am. J. Respir. Crit. Care Med 203, 746–755 (2021).
Won, C. H. J. et al. Sex differences in obstructive sleep apnea phenotypes, the multi-ethnic study of atherosclerosis. Sleep 43, zsz274 (2020).
Maury, G. et al. Mandible behaviour interpretation during wakefulness, sleep and sleep-disordered breathing. J. Sleep. Res 23, 709–716 (2014).
Pépin, J. L., Guillot, M., Tamisier, R. & Lévy, P. The upper airway resistance syndrome. Respiration 83, 559–566 (2012).
Azarbarzin, A., Labarca, G., Kwon, Y. & Wellman, A. Physiologic consequences of upper airway obstruction in sleep apnea. Chest 166, 1209–1217 (2024).
Thapa, R. et al. SleepFM: multi-modal representation learning for sleep across brain activity. ECG and Respiratory Signals, 48019–48037 (2024).
Morris, J. L. et al. Symptom subtype progression in obstructive sleep apnea over 5 years. J. Clin. Sleep Med. 20, 1773–1783 (2024).
Azarbarzin, A. et al. The hypoxic burden of sleep apnoea predicts cardiovascular disease-related mortality: the Osteoporotic Fractures in Men Study and the Sleep Heart Health Study. Eur. Heart J. 40, 1149–1157 (2019).
Nasiri, S. et al. CAISR: achieving human-level performance in automated sleep analysis across all clinical sleep metrics. Sleep 48, zsaf134 (2025).
Stephansen, J. B. et al. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat. Commun. 9, 5229 (2018).
Carvelli, L. et al. Design of a deep learning model for automatic scoring of periodic and non-periodic leg movements during sleep validated against multiple human experts. Sleep. Med 69, 109–119 (2020).
Aittokallio, T., Nevalainen, O., Pursiheimo, U., Saaresranta, T. & Polo, O. Classification of nasal inspiratory flow shapes by attributed finite automata. Comput. Biomed. Res. 32, 34–55 (1999).
Aittokallio, T., Malminen, J. S., Pahikkala, T., Polo, O. & Nevalainen, O. S. Inspiratory flow shape clustering: an automated method to monitor upper airway performance during sleep. Comput Methods Prog. Biomed. 85, 8–18 (2007).
Cohen, O. et al. The great controversy of obstructive sleep apnea treatment for cardiovascular risk benefit: advancing the science through expert consensus. An official American Thoracic Society workshop report. Ann. Am. Thorac. Soc. https://doi.org/10.1513/ANNALSATS.202409-981ST (2024).
Pépin, J. L. et al. Multidimensional phenotyping to distinguish among central (CSA), obstructive (OSA) and co-existing central and obstructive sleep apnea (CSA-OSA) phenotypes in real-world data. Sleep. Med 124, 426–433 (2024).
Chen, X. et al. Racial/Ethnic differences in sleep disturbances: the Multi-Ethnic Study of Atherosclerosis (MESA). Sleep 38, 877–888 (2015).
Blackwell, T. et al. Associations between sleep architecture and sleep-disordered breathing and cognition in older community-dwelling men: the osteoporotic fractures in men sleep study. J. Am. Geriatr. Soc. 59, 2217–2225 (2011).
Young, T. et al. Burden of sleep apnea: rationale, design, and major findings of the Wisconsin Sleep Cohort Study. WMJ 108, 246 (2009).
Redline, S. et al. The familial aggregation of obstructive sleep apnea. Am. J. Respir. Crit. Care Med 151, 682–687 (1995).
Chambon, S., Thorey, V., Arnal, P. J., Mignot, E. & Gramfort, A. DOSED: A deep learning approach to detect multiple sleep micro-events in EEG signal. J. Neurosci. Methods 321, 64–78 (2019).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: unified, real-time object detection. in 779–788 (2016).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. in 770–778 (2016).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. in 448–456 (PMLR, 2015).
Nair, V. & Hinton, G. E. Rectified linear units improve restricted Boltzmann machines. Proceedings of the 27th International Conference on Machine Learning (ICML). 807–814 (2010).
Srivastava, N., Hinton, G., Krizhevsky, A. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Huber, P. J. Robust estimation of a location parameter. Ann. Math. Stat. 35, 73–101 (1964).
Kingma, D. P. & Ba, J. L. Adam: a method for stochastic optimization. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. https://arxiv.org/abs/1412.6980v9 (2014).
Acknowledgments
This research was funded by Stanford University, Mignot Lab, Danish Center for Sleep Medicine, and the Technical University of Denmark. M.R.K. was awarded with Stibo, Augustinus, Knud Højgaard, Otto Mønsted, William Demant, Director Einar Hansen’s and wife Mrs. Vera Hansen’s, DDSA, Viet-Jacobsen, Vera and Carl Johan Michaelsen, Marie and M.B. Richters, Idella, and Rienholdt W. Jorck and Wifes foundations. NIH NHLBI (R01HL146697) funded S.S. The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128.The National Heart, Lung, and Blood Institute (NHLBI) provides funding for the MrOS Sleep ancillary study “Outcomes of Sleep Disorders in Older Men” under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838, and R01 HL070839. The Multi-Ethnic Study of Atherosclerosis (MESA) Sleep Ancillary study was funded by NIH-NHLBI Association of Sleep Disorders with Cardiovascular Health Across Ethnic Groups (RO1 HL098433). MESA is supported by NHLBI-funded contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168 and N01-HC-95169 from the National Heart, Lung, and Blood Institute, and by cooperative agreements UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420 funded by NCATS. The National Sleep Research Resource was supported by the National Heart, Lung, and Blood Institute (R24 HL114473, 75N92019R002). The Cleveland Family Study (CFS) was supported by grants from the National Institutes of Health (HL46380, M01 RR00080-39, T32-HL07567, RO1-46380). The National Sleep Research Resource was supported by the National Heart, Lung, and Blood Institute (R24 HL114473, 75N92019R002). We would like to acknowledge Helge B. D. Sorensen’s contribution to this work, who passed before the work was submitted. We would like to acknowledge Jan Ruud Hansen for his assistance in making the linear regression analysis of ABED AHI detection performance.
Author information
Authors and Affiliations
Contributions
M.R.K. laid out the design of the study, conducted the analyses, wrote the source code for the project from preprocessing to evaluation and analyses, and optimized the deep learning architecture and objectives in relation to sleep apnea and implemented the stepwise linear regression and apnotyping. U.H. assisted in writing, optimized the deep learning architecture and learning objectives in relation to sleep apnea detection, and supervised the analyses. A.B. provided wake and arousal probabilities for all cohorts, assisted in the optimization of deep learning and objectives along with the apnotyping, and supervised the analyses. M.O. assisted in creating figures based on analyses. S.S. and S.R. contributed datasets and provided the loop gain, arousal threshold, and pharyngeal muscle compensation. K.L.S. contributed data. P.J. participated in the design of the study. E.M. participated in the design of the study, supervised the analyses and the writing. O.S. supervised the analyses. O.C. reviewed the signal-processing related issues. All authors contributed to manuscript writing and helped revise the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.S. received grant support from Apnimed, Prosomnus, and Dynaflex and has served as a consultant for Apnimed, Nox Medical, Inspire Medical Systems, Eli Lilly, Respicardia, LinguaFlex, Forepont, and Achaemenid. He receives royalties for intellectual property pertaining to combination pharmacotherapy for sleep apnea via his Institution. He is a co-inventor of intellectual property pertaining to wearable sleep apnea phenotyping, also via his Institution. His industry interactions are actively managed by his Institution. S.R. received consulting fees from Eli Lilly (not related to the project) and funding from NIH. E.M. received a grant from the ResMed Foundation to study questionnaire predictors of SDB, unrelated to this work. U.H. is an employee of BioSerenity. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kjaer, M.R., Hanif, U., Brink-Kjaer, A. et al. Expert-level probabilistic breathing event detector informs phenotyping of sleep apnea. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69163-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69163-z