Abstract
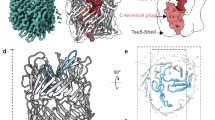
Horizontal gene transfer events were crucial in the emergence of multicellular life. A striking example is the acquisition of Teneurins, putative surface-exposed toxins in bacteria that function as cell adhesion receptors in metazoan neuronal development. Here, we demonstrate the evolutionary relationships between metazoan and bacterial Teneurins. We use cryogenic electron microscopy and bioinformatic analysis to show that bacterial Teneurins harbour a toxic protein in a proteinaceous shell. They are rare but widely distributed across bacterial taxa and are predominantly seen in species with complex social behaviours, suggesting roles in cell-to-cell interaction. This work confirms that metazoan Teneurins are repurposed bacterial toxins that have evolved to be essential mediators of intercellular communication in all advanced nervous systems. Their acquisition was a key event in the evolution of metazoans.
Similar content being viewed by others
Data availability
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-52847 (BiTLP); and EMD-71831 (BiTLPFL). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes 9IFO (BiTLP); and 9PT5 (BiTLPFL). The source data underlying Figs. 2, 3b and e, 4d and Supplementary Fig. 6c are provide as Source Data files. Unedited gels and Western blots are shown in Supplementary Fig. 8. Molecular dynamics trajectories are available from the corresponding authors upon request. Source data are provided with this paper.
References
Tucker, R. P., Beckmann, J., Leachman, N. T., Schöler, J. & Chiquet-Ehrismann, R. Phylogenetic analysis of the teneurins: conserved features and premetazoan ancestry. Mol. Biol. Evol. 29, 1019–1029 (2012).
Wides, R. The natural history of teneurins: a billion years of evolution in three key steps. Front. Neurosci. 13 (2019).
Jackson, V. A., Busby, J. N., Janssen, B. J. C., Lott, J. S. & Seiradake, E. Teneurin structures are composed of ancient bacterial protein domains. Frontiers in Neuroscience 13 (2019).
del Toro, D. et al. Structural basis of teneurin-latrophilin interaction in repulsive guidance of migrating neurons. Cell 180, 323–339.e19 (2020).
Cortés, E., Pak, J. S. & Özkan, E. Structure and evolution of neuronal wiring receptors and ligands. Dev. Dyn. 252, 27–60 (2023).
Lovejoy, D. A. & Pavlović, T. Role of the teneurins, teneurin C-terminal associated peptides (TCAP) in reproduction: clinical perspectives. Horm. Mol. Biol. Clin. Investig. 24, 83–90 (2015).
Burbach, J. P. H. & Meijer, D. H. Latrophilin’s social protein network. Front Neurosci. 13, 643 (2019).
Tucker, R. P. Teneurins: domain architecture, evolutionary origins, and patterns of expression. Front. Neurosci. 12 (2018).
Cai, K., Zhang, X. & Bai, X. Cryo-electron microscopic analysis of single-pass transmembrane receptors. Chem. Rev. 122, 13952–13988 (2022).
Gogou, C. et al. Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition. Nat. Commun. 15, 3648 (2024).
Mosca, T. J., Hong, W., Dani, V. S., Favaloro, V. & Luo, L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237–241 (2012).
Suzuki, N. et al. Teneurin-4 is a novel regulator of oligodendrocyte differentiation and myelination of small-diameter axons in the CNS. J. Neurosci. 32, 11586–11599 (2012).
Berns, D. S., DeNardo, L. A., Pederick, D. T. & Luo, L. Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature 554, 328–333 (2018).
Sando, R., Jiang, X. & Südhof, T. C. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363, eaav7969 (2019).
Pederick, D. T. et al. Reciprocal repulsions instruct the precise assembly of parallel hippocampal networks. Science 372, 1068–1073 (2021).
Mosca, T. J. On the Teneurin track: a new synaptic organization molecule emerges. Front. Cell. Neurosci. 9, 204 (2015).
Alkelai, A. et al. A role for TENM1 mutations in congenital general anosmia. Clin. Genet. 90, 211–219 (2016).
Hor, H. et al. Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum. Mol. Genet. 24, 5677 (2015).
Pu, J.-L. et al. Parkinson’s disease in teneurin transmembrane protein 4 (TENM4) Mutation Carriers. Front Genet 11, 598064 (2020).
Li, S., DeLisi, L. E. & McDonough, S. I. Rare germline variants in individuals diagnosed with schizophrenia within multiplex families. Psychiatry Res. 303, 114038 (2021).
Heinrich, A. et al. The risk variant in ODZ4 for bipolar disorder impacts on amygdala activation during reward processing. Bipolar Disord. 15, 440–445 (2013).
Xu, C. et al. Molecular and cellular mechanisms of teneurin signaling in synaptic partner matching. Cell 187, 5081–5101.e19 (2024).
Peppino, G. et al. Teneurins: role in cancer and potential role as diagnostic biomarkers and targets for therapy. Int J. Mol. Sci. 22, 2321 (2021).
Jackson, V. A. et al. Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Nat. Commun. 9, 1079 (2018).
Li, J. et al. Structural basis for teneurin function in circuit-wiring: a toxin motif at the synapse. Cell 173, 735–748.e15 (2018).
Li, J. et al. Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism. Nat. Commun. 11, 2140 (2020).
Meijer, D. H., Frias, C. P., Beugelink, J. W., Deurloo, Y. N. & Janssen, B. J. C. Teneurin4 dimer structures reveal a calcium-stabilized compact conformation supporting homomeric trans-interactions. EMBO J. 41, e107505 (2022).
Li, J., Bandekar, S. J. & Araç, D. The structure of fly Teneurin-m reveals an asymmetric self-assembly that allows expansion into zippers. EMBO Rep. 24, e56728 (2023).
Busby, J. N., Panjikar, S., Landsberg, M. J., Hurst, M. R. H. & Lott, J. S. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501, 547–550 (2013).
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M. & Aravind, L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012).
Ferralli, J., Tucker, R. P. & Chiquet-Ehrismann, R. The teneurin C-terminal domain possesses nuclease activity and is apoptogenic. Biol. Open 7, bio031765 (2018).
Jurėnas, D. & Journet, L. Activity, delivery, and diversity of Type VI secretion effectors. Mol. Microbiol. 115, 383–394 (2021).
Koskiniemi, S. et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. 110, 7032–7037 (2013).
Pei, T.-T. et al. Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 11, 1865 (2020).
Günther, P. et al. Structure of a bacterial Rhs effector exported by the type VI secretion system. PLOS Pathog. 18, e1010182 (2022).
Tang, L. et al. Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP. Cell Rep. 41, 111732 (2022).
González-Magaña, A. et al. Structural and functional insights into the delivery of a bacterial Rhs pore-forming toxin to the membrane. Nat. Commun. 14, 7808 (2023).
Hernandez, R. E., Gallegos-Monterrosa, R. & Coulthurst, S. J. Type VI secretion system effector proteins: Effective weapons for bacterial competitiveness. Cell. Microbiol. 22, e13241 (2020).
Allsopp, L. P. & Bernal, P. Killing in the name of: T6SS structure and effector diversity. Microbiology 169, 001367 (2023).
Roderer, D., Schubert, E., Sitsel, O. & Raunser, S. Towards the application of Tc toxins as a universal protein translocation system. Nat. Commun. 10, 5263 (2019).
Aleksandrova, N. A., Roche, S. G., Low, Y. S. & Landsberg, M. J. Recent insights into mechanisms of cellular toxicity and cell recognition associated with the ABC family of pore-forming toxins. Biochem. Soc. Trans. 51, 1235–1244 (2023).
Minet, A. D. & Chiquet-Ehrismann, R. Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene 257, 87–97 (2000).
Tucker, R. P. & Chiquet-Ehrismann, R. Teneurins: A conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol. 290, 237–245 (2006).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124 (2018).
Fagan, R. P. & Fairweather, N. F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol 12, 211–222 (2014).
Ravi, J. & Fioravanti, A. S-layers: The Proteinaceous Multifunctional Armors of Gram-Positive Pathogens. Front. Microbiol. 12, (2021).
Green, E. R. & Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol Spectr. 4, 4.1.13 (2016).
Luo, Y. et al. The love and hate relationship between T5SS and other secretion systems in bacteria. Int. J. Mol. Sci. 25, 281 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Rooney, A. P., Price, N. P. J., Ehrhardt, C., Swezey, J. L. & Bannan, J. D. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evolut. Microbiol. 59, 2429–2436 (2009).
Dunlap, C. A., Bowman, M. J. & Zeigler, D. R. Promotion of Bacillus subtilis subsp. inaquosorum, Bacillus subtilis subsp. spizizenii and Bacillus subtilis subsp. stercoris to species status. Antonie van. Leeuwenhoek 113, 1–12 (2020).
Jackson, A. P., Thomas, G. H., Parkhill, J. & Thomson, N. R. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10, 584 (2009).
Jurėnas, D. et al. Mounting, structure and autocleavage of a type VI secretion-associated Rhs polymorphic toxin. Nat. Commun. 12, 6998 (2021).
Hayes, B. K. et al. Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery. Nat. Commun. 15, 8709 (2024).
Meusch, D. et al. Mechanism of Tc toxin action revealed in molecular detail. Nature 508, 61–65 (2014).
Kielkopf, C. S., Shneider, M. M., Leiman, P. G. & Taylor, N. M. I. T6SS-associated Rhs toxin-encapsulating shells: Structural and bioinformatical insights into bacterial weaponry and self-protection. Structure 0, (2024).
Ma, J. et al. PAAR-Rhs proteins harbor various C-terminal toxins to diversify the antibacterial pathways of type VI secretion systems. Environ. Microbiol. 19, 345–360 (2017).
Ruhe, Z. C., Low, D. A. & Hayes, C. S. Polymorphic toxins and their immunity proteins: diversity, evolution and mechanisms of delivery. Annu Rev. Microbiol 74, 497–520 (2020).
Song, N. et al. Genome-wide dissection reveals diverse pathogenic roles of bacterial Tc toxins. PLOS Pathog. 17, e1009102 (2021).
Weiss, M. S., Blanke, S. R., Collier, R. J. & Eisenberg, D. Structure of the isolated catalytic domain of diphtheria toxin. Biochemistry 34, 773–781 (1995).
Lyons, B. et al. Scabin, a Novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies*. J. Biol. Chem. 291, 11198–11215 (2016).
Mikolčević, P., Hloušek-Kasun, A., Ahel, I. & Mikoč, A. ADP-ribosylation systems in bacteria and viruses. Comput Struct. Biotechnol. J. 19, 2366–2383 (2021).
Baumgartner, S. & Wides, R. Discovery of teneurins. Front Neurosci. 13, 230 (2019).
Jamet, A. & Nassif, X. New Players in the Toxin Field: Polymorphic Toxin Systems in Bacteria. mBio 6, https://doi.org/10.1128/mbio.00285-15 (2015).
Li, H., Tan, Y. & Zhang, D. Genomic discovery and structural dissection of a novel type of polymorphic toxin system in gram-positive bacteria. Computational Struct. Biotechnol. J. 20, 4517–4531 (2022).
Livingstone, P. G., Morphew, R. M. & Whitworth, D. E. Genome Sequencing and Pan-Genome Analysis of 23 Corallococcus spp. Strains Reveal Unexpected Diversity, With Particular Plasticity of Predatory Gene Sets. Front. Microbiol. 9, (2018).
Livingstone, P. G. et al. Predatory Organisms with Untapped Biosynthetic Potential: Descriptions of Novel Corallococcus Species C. aberystwythensis sp. nov., C. carmarthensis sp. nov., C. exercitus sp. nov., C. interemptor sp. nov., C. llansteffanensis sp. nov., C. praedator sp. nov., C. sicarius sp. nov., and C. terminator sp. nov. Appl. Environ. Microbiol. 86, e01931 (2020).
Sitsel, O. et al. Yersinia entomophaga Tc toxin is released by T10SS-dependent lysis of specialized cell subpopulations. Nat. Microbiol 9, 390–404 (2024).
Gatsogiannis, C. et al. Tc toxin activation requires unfolding and refolding of a β-propeller. Nature 563, 209–213 (2018).
Youderian, P. & Hartzell, P. L. Triple mutants uncover three new genes required for social motility in myxococcus xanthus. Genetics 177, 557–566 (2007).
Sirota-Madi, A. et al. Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics 11, 710 (2010).
Muñoz-Dorado, J., Marcos-Torres, F. J., García-Bravo, E., Moraleda-Muñoz, A. & Pérez, J. Myxobacteria: Moving, Killing, Feeding, and Surviving Together. Front. Microbiol. 7, (2016).
Sita, L. V., Diniz, G. B., Horta-Junior, J. A. C., Casatti, C. A. & Bittencourt, J. C. Nomenclature and Comparative Morphology of the Teneurin/TCAP/ADGRL Protein Families. Front. Neurosci. 13 (2019).
Yue, J., Sun, G., Hu, X. & Huang, J. The scale and evolutionary significance of horizontal gene transfer in the choanoflagellate Monosiga brevicollis. BMC Genomics 14, 729 (2013).
Kiørboe, T. Predation in a Microbial World: Mechanisms and Trade-Offs of Flagellate Foraging. Annu. Rev. Mar. Sci. 16, 361–381 (2024).
Tucker, R. P. Horizontal Gene Transfer in Choanoflagellates. J. Exp. Zool. Part B: Mol. Dev. Evol. 320, 1–9 (2013).
Beckmann, J., Schubert, R., Chiquet-Ehrismann, R. & Müller, D. J. Deciphering teneurin domains that facilitate cellular recognition, cell–cell adhesion, and neurite outgrowth using atomic force microscopy-based single-cell force spectroscopy. Nano Lett. 13, 2937–2946 (2013).
Silva, J.-P. et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl. Acad. Sci. USA 108, 12113–12118 (2011).
Ram, A. & Lo, A. W. Is smaller better? A proposal to use bacteria for neuroscientific modeling. Front Comput Neurosci. 12, 7 (2018).
Prindle, A. et al. Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63 (2015).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
HMMER. http://hmmer.org/.
Troshin, P. V., Procter, J. B. & Barton, G. J. Java bioinformatics analysis web services for multiple sequence alignment—JABAWS:MSA. Bioinformatics 27, 2001–2002 (2011).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–W469 (2008).
Lemoine, F. et al. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 47, W260–W265 (2019).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Almagro Armenteros, J. J. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019).
Wang, J. et al. The conserved domain database in 2023. Nucleic Acids Res 51, D384–D388 (2023).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49, D344–D354 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Romier, C. et al. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Cryst. D. 62, 1232–1242 (2006).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Mastronarde, D. N. SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microanal 9, 1182–1183 (2003).
Caesar, J. et al. SIMPLE 3.0. Stream single-particle cryo-EM analysis in real time. J. Struct. Biol.: X 4, 100040 (2020).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Cryst. D. 75, 861–877 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Prisant, M. G., Williams, C. J., Chen, V. B., Richardson, J. S. & Richardson, D. C. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink “waters,” and NGL Viewer to recapture online 3D graphics. Protein Sci. 29, 315–329 (2020).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Kidmose, R. T. et al. Namdinator – automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Cryst. D. 74, 519–530 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Kroon, P. C. et al. Martinize2 and Vermouth: Unified Framework for Topology Generation. eLife 12, (2024).
Wassenaar, T. A., Ingólfsson, H. I., Böckmann, R. A., Tieleman, D. P. & Marrink, S. J. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015).
Souza, P. C. T. et al. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Tironi, I. G., Sperb, R., Smith, P. E. & van Gunsteren, W. F. A generalized reaction field method for molecular dynamics simulations. J. Chem. Phys. 102, 5451–5459 (1995).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Computational Chem. 18, 1463–1472 (1997).
Jackson, V. et al. The guidance and adhesion protein FLRT2 dimerizes in cis via dual small-X3-small transmembrane motifs. Structure 30, 1354–1365.e5 (2022).
Cornet, J. et al. There and back again: bridging meso- and nano-scales to understand lipid vesicle patterning. Soft Matter 20, 4998–5013 (2024).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol 1, 1–7 (2016).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of Image Analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We acknowledge the Central Oxford Structural Molecular Imaging Center (COSMIC) electron microscopy where the cryo-EM data of BiTLP was collected. We also thank Dr Joseph Caesar for support with data processing. We would also like to acknowledge Australian Microscopy and Microanalysis Research Facility at the Center for Microscopy and Microanalysis located at the University of Queensland where the data for BiTLPFL is collected. We also thank Professor Steve Kelly for his valuable help and advice on the phylogenetic tree inference. We also give thanks to the Fribourg Lab for supplying the petMCN vector used to clone Bacillus inaquosorum TLP, and to Dr. Valentine Lagage and Dr. Diviya Choudhary for providing the pBAD vector used to clone all bacterial TLP CTD and immunity genes. We finally thank the Dean lab for providing us with the Alphafold model of Bacillus inaquosorum TLP. F.R. was funded by the Browne research fellowship, the Queen’s college Oxford. J.B. is funded by a Wellcome Trust Biomedical Resource Grant (number 218205/Z/19/Z, PubMLST: Disseminating and exploiting bacterial diversity data for public health benefit). Research in the E.S. lab was supported by the Wellcome Trust (202827/Z/16/Z and 226647/Z/22/Z) and the EMBO Young Investigator Programme.
Author information
Authors and Affiliations
Contributions
F.R. and E.S. conceptualized the study and designed the experiments. F.R. and C.M.R. performed the biochemical experiments. J.C.Z. and Y.S.L. performed the structural biology experiments. J.C.Z. and L.B. established and implemented the cryo-EM methodology in the laboratory. F.R., E.S., K.E.O., and E.L. built and refined the BiTLP model, while Y.S.L. and M.J.L. built and refined the BiTLPFL model. J.B. performed the bioinformatic analyses. J.S. performed the microscopy experiments. A.S. and M.C. performed the structural predictions and molecular dynamics simulations. F.R., J.S., A.S., J.C.Z. and J.B. analysed the data. F.R., J.S., A.S., J.B., C.M.R., Y.S.L., M.J.L., J.S.L., C.K., M.C., M.C.J.M., and E.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Raoelijaona, F., Szczepaniak, J., Schahl, A. et al. Ancestral neuronal receptors are bacterial accessory toxins. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69246-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69246-x