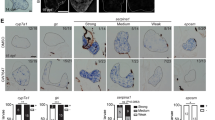
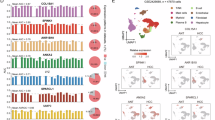
Abstract
Colorectal cancer liver metastasis (CRLM) is a leading cause of mortality, driven by poorly defined molecular interactions within the hepatic niche. Here, we identify a distinct population of pro-metastatic Early Growth Response 1 (Egr1)+ neutrophils that accumulate in the pre-metastatic liver. Mechanistically, we show that KIAA1199-high cancer cells secrete granulin-rich extracellular vesicles, which are internalized by hepatocytes. This uptake triggers a subset of functionally reprogrammed hepatocytes, characterized by a profound metabolic reprogramming and the suppression of peroxisome proliferator-activated receptor gamma (PPARγ) signaling, leading to increased secretion of Serum Amyloid A2 (SAA2). Hepatocyte-derived SAA2 subsequently activates Formyl Peptide Receptor 2 (FPR2) on neutrophils, stabilizing Egr1-driven transcriptional program via the PI3K-AKT pathway to enhance neutrophil survival and pro-angiogenic activity. These Egr1+ neutrophils co-localize with reprogrammed hepatocytes at the tumor-liver interface, where they promote vascular remodeling to facilitate metastatic colonization. Pharmacological restoration of PPARγ or FPR2 inhibition abrogate CRLM in preclinical models in female mice. Furthermore, a combined KIAA1199-SAA2 signature predicts liver metastasis risk in patients. Our findings delineate a KIAA1199-PPARγ/SAA2-Egr1 axis orchestrating the pre-metastatic niche and propose metabolic normalization as a preventative strategy for liver metastasis.
Similar content being viewed by others
Data availability
The publicly available data used in this study are available in the Gene Expression Omnibus (GEO) under accession code GSE225857, the National Omics Data Encyclopedia (NODE) under accession code OEP00001756, and The Cancer Genome Atlas (TCGA) Pan-Cancer dataset (https://www.cancer.gov/tcga). The single-cell RNA-sequencing data generated in this study have been deposited in the Genome Sequence Archive (GSA) under the accession code PRJCA046174. The proteomics and transcriptomics data generated in this study have been deposited in the iProX database under accession code IPX0014830000 and are also available at Zenodo under https://doi.org/10.5281/zenodo.18205510. The remaining data are available within the Article, Supplementary Information, or Source Data file, and/or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
All data analyses and processing were performed using publicly available software and established packages, as detailed and cited in the Methods section. The code used in this study has been deposited in Zenodo under https://doi.org/10.5281/zenodo.18205510.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. Cancer J. Clinicians 72, 7–33 (2022).
Zheng, R. S. et al. Cancer incidence and mortality in China, 2022]. Zhonghua Zhong Liu Za Zhi 46, 221–231 (2024).
Raphael, M. J. & Karanicolas, P. J. Regional Therapy for colorectal cancer liver metastases: which modality and when? J. Clin. Oncol. 40, 2806–2817 (2022).
Datta, J., Narayan, R. R., Kemeny, N. E. & D’Angelic, M. I. Role of hepatic artery infusion chemotherapy in treatment of initially unresectable colorectal liver metastases: a review. JAMA Surg. 154, 768–776 (2019).
Patras, L., Shaashua, L., Matei, I. & Lyden, D. Immune determinants of the pre-metastatic niche. Cancer Cell 41, 546–572 (2023).
Liu, Y. & Cao, X. Characteristics and significance of the pre-metastatic niche. Cancer Cell 30, 668–681 (2016).
Wang, Y. et al. Pre-metastatic niche: formation, characteristics and therapeutic implication. Signal Transduct. Target Ther. 9, 236 (2024).
Zeng, Z. et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 9, 5395 (2018).
Babak, M. V., Zalutsky, M. R. & Balyasnikova, I. V. Heterogeneity and vascular permeability of breast cancer brain metastases. Cancer Lett. 489, 174–181 (2020).
Coffelt, S. B., Wellenstein, M. D. & Visser, K. E. d. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 (2016).
Park, J. et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci. Transl. Med. 8, 361ra138 (2016).
Mantovani, A., Cassatella, M. A., Costantini, C. & Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11, 519–531 (2011).
Hedrick, C. C. & Malanchi, I. Neutrophils in cancer: heterogeneous and multifaceted. Nat. Rev. Immunol. 22, 173–187 (2022).
Christoffersson, G. et al. VEGF-A recruits a proangiogenic MMP-9-delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood 120, 4653–4662 (2012).
Yang, L. et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 583, 133–138 (2020).
Liu, J., Yan, W., Han, P. & Tian, D. The emerging role of KIAA1199 in cancer development and therapy. Biomed. Pharmacother. Biomed. Pharmacotherapie 138, 111507 (2021).
Chen, Y. et al. The role of CEMIP in tumors: an update based on cellular and molecular insights. Biomed. Pharmacother. 146, 112504 (2022).
Li, L. et al. Targeting pyroptosis reverses KIAA1199-mediated immunotherapy resistance in colorectal cancer. J Immunother. Cancer 13, e010000 (2025).
Zhao, L. et al. KIAA1199 promotes metastasis of colorectal cancer cells via microtubule destabilization regulated by a PP2A/stathmin pathway. Oncogene 38, 935–949 (2019).
Zhang, D. et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J. Cancer 140, 2298–2309 (2017).
Hua, Q. et al. CEMIP, a novel adaptor protein of OGT, promotes colorectal cancer metastasis through glutamine metabolic reprogramming via reciprocal regulation of β-catenin. Oncogene 40, 6443–6455 (2021).
Wang, H. et al. KIAA1199 drives immune suppression to promote colorectal cancer liver metastasis by modulating neutrophil infiltration. Hepatology 76, 967–981 (2022).
Zhang, B. et al. Tumor CEMIP drives immune evasion of colorectal cancer via MHC-I internalization and degradation. J. Immunother. Cancer 11, e005592 (2023).
Xu, G. et al. CEMIP, acting as a scaffold protein for bridging GRAF1 and MIB1, promotes colorectal cancer metastasis via activating CDC42/MAPK pathway. Cell Death Dis. 14, 167 (2023).
Che, L. H. et al. A single-cell atlas of liver metastases of colorectal cancer reveals reprogramming of the tumor microenvironment in response to preoperative chemotherapy. Cell Discov. 7, 80 (2021).
Wang, F. et al. Single-cell and spatial transcriptome analysis reveals the cellular heterogeneity of liver metastatic colorectal cancer. Sci. Adv. 9, eadf5464 (2023).
Wang, B. et al. The role of the transcription factor EGR1 in cancer. Front Oncol. 11, 642547 (2021).
Ji, R. C. Hypoxia and lymphangiogenesis in tumor microenvironment and metastasis. Cancer Lett. 346, 6–16 (2014).
Cheng, J. C., Chang, H. M. & Leung, P. C. Egr-1 mediates epidermal growth factor-induced downregulation of E-cadherin expression via Slug in human ovarian cancer cells. Oncogene 32, 1041–1049 (2013).
Grieshaber-Bouyer, R. et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat. Commun. 12, 2856 (2021).
Pettinella, F. et al. Surface CD52, CD84, and PTGER2 mark mature PMN-MDSCs from cancer patients and G-CSF-treated donors. Cell Rep. Med. 5, 101380 (2024).
Hori, S. FOXP3 as a master regulator of T(reg) cells. Nat. Rev. Immunol. 21, 618–619 (2021).
Bui, T. M. et al. Tissue-specific reprogramming leads to angiogenic neutrophil specialization and tumor vascularization in colorectal cancer. J. Clin. Invest. 134, e174545 (2024).
Wang, H. et al. Neutrophil extracellular traps in homeostasis and disease. Signal Transduct. Target Ther. 9, 235 (2024).
Wu, L. et al. An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression. Cell Res. 33, 585–603 (2023).
Ramirez, C. F. A. & Akkari, L. Myeloid cell path to malignancy: insights into liver cancer. Trends Cancer 11, 591–610 (2025).
Cordoba-Chacon, J. Loss of hepatocyte-specific PPARgamma expression ameliorates early events of steatohepatitis in mice fed the methionine and choline-deficient diet. PPAR Res. 2020, 9735083 (2020).
Francque, S. et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat. Rev. Gastroenterol. Hepatol. 18, 24–39 (2021).
Han, C. Y. et al. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes 56, 2260–2273 (2007).
Lee, J. W. et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature 567, 249–252 (2019).
Harmon, G. S., Lam, M. T. & Glass, C. K. PPARs and lipid ligands in inflammation and metabolism. Chem. Rev. 111, 6321–6340 (2011).
Juurlink, D. N. Rosiglitazone and the case for safety over certainty. JAMA 304, 469–471 (2010).
Ma, Y. et al. Type 2 diabetes and risk of colorectal cancer in two large U.S. prospective cohorts. Br. J. Cancer 119, 1436–1442 (2018).
Peng, H. et al. Hyperglycemia induces an immunosuppressive microenvironment in colorectal cancer liver metastases by recruiting peripheral blood monocytes through the CCL3-CCR1 axis. J. Immunother. Cancer 13, e011310 (2025).
Rada, M., Krzywon, L., Petrillo, S., Lazaris, A. & Metrakos, P. A Retrospective study on the role of metformin in colorectal cancer liver metastases. Biomedicines 11, 731 (2023).
Acknowledgements
The authors thank the patients who participated in this study. This work is supported by the National Natural Science Foundation of China (82272711 to D.J.Z., 82272900 to L.Z., and 82573302, 82373050 to T.Z.).
Author information
Authors and Affiliations
Contributions
L.S.L., K.C., P.Y.Z., G.J.X., and J.G.Z. conducted experiments. L.S.L., K.C., and D.J.Z. wrote the manuscript. L.Z. and D.J.Z. designed research studies and analyzed data. Z.Y.L., D.D.Y., J.Z., P.F.Z., and J.H.R. acquired data. D.J.Z. and Z.T. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Samuel Huber and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, L., Zhao, L., Cao, K. et al. Hepatocytes functionally reprogrammed by KIAA1199-high colorectal cancer cells favour the accumulation of pro-metastatic Egr1+ neutrophils. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69250-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69250-1