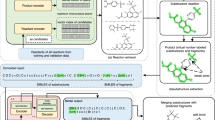
Abstract
Recent advancements in artificial intelligence (AI) have revolutionized the field of 3D molecule generation. However, the lack of effective evaluation methods for 3D conformations limits further improvements. Current techniques, in order to achieve the necessary speed for evaluating large number of AI-generated molecules, often rely on empirical geometric metrics that do not adequately capture various conformational anomalies, or on molecular mechanics energy metrics that exhibit low accuracy and lack atomic or torsional details. To address this gap, we propose a two-stage approach that achieves both high speed and quantum mechanical level accuracy. The first stage, termed the validity test, employs an AI-derived force field to identify atoms with elevated energy resulting from implausible neighboring environments. The second stage, known as the rationality test, utilizes a deep learning network trained on data with density functional theory accuracy to detect rotatable bonds with high torsional energies. To demonstrate the functionality of our evaluation system, we applied our approach to five recently reported 3D molecule generation AI models across 102 targets in Directory of Useful Decoys-Enhanced dataset. To facilitate accessibility for the academic community, our method is available as an open-source package.
Similar content being viewed by others
Data availability
The GM-5K, GM-1K, and DFT-5K datasets generated in this study have been deposited in the Figshare database74 under accession link https://doi.org/10.6084/m9.figshare.27826488.v6. Source data for figures are provided with this paper. Source data are provided with this paper.
Code availability
Our source code for the HEAD and TED models is publicly available on our GitHub repository at https://github.com/stonewiseAIDrugDesign/HEAD_TED. (The code is also included in the Code Ocean capsule21). The code is distributed under MIT license. We obtained the models for Pocket2Mol, TargetDiff, PocketFlow, and PMDM from their official GitHub repositories and used them for molecule generation. For Lingo3DMolv2, we utilized the online service available at https://sw3dmg.stonewise.cn to generate molecules. The service is freely accessible to academic users, and an academic email address is required to receive the generated results.
References
Zeng, X. et al. Deep generative molecular design reshapes drug discovery. Cell Rep. Med. 3, 100794 (2022).
Jiang, Y. et al. PocketFlow is a data-and-knowledge-driven structure-based molecular generative model. Nat. Mach. Intell. 6, 326–337 (2024).
Feng, W. et al. Generation of 3D molecules in pockets via a language model. Nat. Mach. Intell. 6, 62–73 (2024).
Huang, L. et al. A dual diffusion model enables 3D molecule generation and lead optimization based on target pockets. Nat. Commun. 15, 2657 (2024).
Peng, X. et al. Pocket2Mol: Efficient Molecular Sampling Based on 3D Protein Pockets. arXiv:2205.07249 https://ui.adsabs.harvard.edu/abs/2022arXiv220507249P (2022).
Guan, J. et al. 3D Equivariant Diffusion for Target-Aware Molecule Generation and Affinity Prediction. arXiv:2303.03543 https://ui.adsabs.harvard.edu/abs/2023arXiv230303543G (2023).
Wang, L. et al. A pocket-based 3D molecule generative model fueled by experimental electron density. Sci. Rep. 12, 15100 (2022).
Buttenschoen, M., Morris, G. M. & Deane, C. M. PoseBusters: AI-based docking methods fail to generate physically valid poses or generalise to novel sequences. Chem. Sci. 15, 3130–3139 (2024).
Harris, C. et al. Benchmarking Generated Poses: How Rational is Structure-based Drug Design with Generative Models? https://ui.adsabs.harvard.edu/abs/2023arXiv230807413H (2023). arXiv:2308.07413.
Hekkelman, M. L., de Vries, I., Joosten, R. P. & Perrakis, A. J. N. M. AlphaFill. enriching AlphaFold models ligands cofactors. 20, 205–213 (2023).
Ramachandran, S., Kota, P., Ding, F., Dokholyan, N. V. J. P. S., Function, & Bioinformatics. Automated minimization of steric clashes in protein structures. 79, 261-270 (2011).
Kohn, W. & Sham, L. J. J. P. r. Self-consistent equations including exchange and correlation effects. A 140, 1133 (1965).
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
Allinger, N. L., Yuh, Y. H. & Lii, J. H. Molecular mechanics. The MM3 force field for hydrocarbons. 1. J. Am. Chem. Soc. 111, 8551–8566 (1989).
Halgren, T. A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17, 490–519 (1996).
Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Brooks, B. R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Sellers, B. D., James, N. C. & Gobbi, A. A comparison of quantum and molecular mechanical methods to estimate strain energy in druglike fragments. J. Chem. Inf. Model 57, 1265–1275 (2017).
Wang, Y., Walker, B. D., Liu, C. & Ren, P. An efficient approach to large-scale ab initio conformational energy profiles of small molecules. Molecules https://doi.org/10.3390/molecules27238567 (2022).
Mysinger, M. M., Carchia, M., Irwin, J. J. & Shoichet, B. K. Directory of useful decoys, enhanced (dud-e): better ligands and decoys for better benchmarking. J. Medicinal Chem. 55, 6582–6594 (2012).
Fan, F. et al. Assessing conformation validity and rationality of deep learning-generated 3d molecules [source code]. Code Ocean https://doi.org/10.24433/CO.3893415.v2 (2025).
Devereux, C. et al. Extending the applicability of the ani deep learning molecular potential to sulfur and halogens. J. Chem. Theory Comput. 16, 4192–4202 (2020).
Landrum, G. e. a. RDKit: open-source cheminformatics software. GitHub https://github.com/rdkit/rdkit (2016).
Tong, J. & Zhao, S. Large-scale analysis of bioactive ligand conformational strain energy by ab initio calculation. J. Chem. Inf. Model 61, 1180–1192 (2021).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Roos, K. et al. OPLS3e: extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 15, 1863–1874 (2019).
Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. The cambridge structural database. Acta Crystallogr B Struct. Sci. Cryst. Eng. Mater. 72, 171–179 (2016).
Santra, G., Sylvetsky, N. & Martin, J. M. J. T. J. o. P. C. A. Minimally empirical double-hybrid functionals trained against the GMTKN55 database: revDSD-PBEP86-D4, revDOD-PBE-D4, and DOD-SCAN-D4. 123, 5129–5143 (2019).
Hellweg, A. & Rappoport, D. J. P. C. C. P. Development of new auxiliary basis functions of the Karlsruhe segmented contracted basis sets including diffuse basis functions (def2-SVPD, def2-TZVPPD, and def2-QVPPD) for RI-MP2 and RI-CC calculations. 17, 1010–1017 (2015).
Grimme, S., Ehrlich, S. & Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Computational Chem. 32, 1456–1465 (2011).
Genheden, S. & Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461 (2015).
Miller, B. R. et al. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput 8, 3314–3321 (2012).
Cieplinski, T., Danel, T., Podlewska, S. & Jastrzebski, S. Generative models should at least be able to design molecules that dock well: a new benchmark. J. Chem. Inf. Model 63, 3238–3247 (2023).
Jocys, Z., Grundy, J. & Farrahi, K. DrugPose: benchmarking 3D generative methods for early stage drug discovery. Digital Discov. 3, 1308–1318 (2024).
Koes, D. R., Baumgartner, M. P. & Camacho, C. J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model 53, 1893–1904 (2013).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform 1, 8 (2009).
Rai, B. K. et al. TorsionNet: a deep neural network to rapidly predict small-molecule torsional energy profiles with the accuracy of quantum mechanics. J. Chem. Inf. Modeling 62, 785–800 (2022).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46, D1074–D1082 (2018).
Clark, C. G. et al. Structure based design of macrocyclic factor XIa inhibitors: Discovery of cyclic P1 linker moieties with improved oral bioavailability. Bioorg. Med Chem. Lett. 29, 126604 (2019).
Kovács, D. P. et al. MACE-OFF: short-range transferable machine learning force fields for organic molecules. J. Am. Chem. Soc. 147, 17598–17611 (2025).
Eastman, P. et al. SPICE, A Dataset of Drug-like Molecules and Peptides for Training Machine Learning Potentials. Sci. Data 10, 11 (2023).
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput Aided Mol. Des. 27, 221–234 (2013).
Wallace, E. R. S., Frey, N. C. & Rackers, J. A. Strain problems got you in a twist? try strainrelief: a quantum-accurate tool for ligand strain calculations. J. Chem. Inf. Model 65, 6613–6620 (2025).
Watts, K. S. et al. ConfGen: a conformational search method for efficient generation of bioactive conformers. J. Chem. Inf. Modeling 50, 534–546 (2010).
Peach, M. L., Cachau, R. E. & Nicklaus, M. C. Conformational energy range of ligands in protein crystal structures: The difficult quest for accurate understanding. J. Mol. Recognit. https://doi.org/10.1002/jmr.2618 (2017).
Brameld, K. A., Kuhn, B., Reuter, D. C. & Stahl, M. Small molecule conformational preferences derived from crystal structure data. A medicinal chemistry focused analysis. J. Chem. Inf. Model 48, 1–24 (2008).
Zhao, L., Pu, M., Wang, H., Ma, X. & Zhang, Y. J. Modified electrostatic complementary score function and its application boundary exploration in drug design. J. Chem. Inf. Model 62, 4420–4426 (2022).
Ding, K. et al. Observing noncovalent interactions in experimental electron density for macromolecular systems: a novel perspective for protein-ligand interaction research. J. Chem. Inf. Model 62, 1734–1743 (2022).
Unke, O. T. et al. Machine learning force fields. Chem. Rev. 121, 10142–10186 (2021).
Kocer, E., Ko, T. W. & Behler, J. Neural network potentials: a concise overview of methods. Annu Rev. Phys. Chem. 73, 163–186 (2022).
Wu, S. et al. Applications and advances in machine learning force fields. J. Chem. Inf. Model 63, 6972–6985 (2023).
Zhang, L. et al. Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics. Phys. Rev. Lett. 120, 143001 (2018).
Schutt, K. T., Sauceda, H. E., Kindermans, P. J., Tkatchenko, A. & Muller, K. R. SchNet - A deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Fu, W. et al. Enhancing molecular energy predictions with physically constrained modifications to the neural network potential. J. Chem. Theory Comput 20, 4533–4544 (2024).
Smith, J. S. et al. Approaching coupled cluster accuracy with a general-purpose neural network potential through transfer learning. Nat. Commun. 10, 2903 (2019).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Behler, J. Representing potential energy surfaces by high-dimensional neural network potentials. J. Phys. Condens Matter 26, 183001 (2014).
Zubatyuk, R., Smith, J. S., Leszczynski, J. & Isayev, O. Accurate and transferable multitask prediction of chemical properties with an atoms-in-molecules neural network. Sci. Adv. 5, eaav6490 (2019).
Gasteiger, J., Groß, J. & Günnemann, S. J. a. e.-p. Directional Message Passing for Molecular Graphs. arXiv:2003.03123. https://ui.adsabs.harvard.edu/abs/2020arXiv200303123G (2020).
Batatia, I. et al. The Design Space of E(3)-Equivariant Atom-Centered Interatomic Potentials. arXiv:2205.06643. https://ui.adsabs.harvard.edu/abs/2022arXiv220506643B (2022).
Ramakrishnan, R., Dral, P. O., Rupp, M. & von Lilienfeld, O. A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014).
Qiao, Z., Welborn, M., Anandkumar, A., Manby, F. R. & Miller, T. F. 3rd. OrbNet: Deep learning for quantum chemistry using symmetry-adapted atomic-orbital features. J. Chem. Phys. 153, 124111 (2020).
Isert, C., Atz, K., Jimenez-Luna, J. & Schneider, G. QMugs, quantum mechanical properties of drug-like molecules. Sci. Data 9, 273 (2022).
Rai, B. K. et al. Comprehensive assessment of torsional strain in crystal structures of small molecules and protein-ligand complexes using ab initio calculations. J. Chem. Inf. Model 59, 4195–4208 (2019).
Gale, J. D., LeBlanc, L. M., Spackman, P. R., Silvestri, A. & Raiteri, P. A universal force field for materials, periodic gfn-ff: implementation and examination. J. Chem. Theory Comput. 17, 7827–7849 (2021).
Spicher, S. & Grimme, S. Robust atomistic modeling of materials, organometallic, and biochemical systems. Angew. Chem. Int Ed. Engl. 59, 15665–15673 (2020).
Neese, F., Wennmohs, F., Becker, U. & Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 152, 224108 (2020).
Bajusz, D., Racz, A. & Heberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform 7, 20 (2015).
Vaswani, A. et al. Attention Is All You Need. arXiv:1706.03762. https://ui.adsabs.harvard.edu/abs/2017arXiv170603762V (2017).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713 (2015).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph Model 25, 247–260 (2006).
Fan, F. et al. Data for HEAD_TED. Figshare https://doi.org/10.6084/m9.figshare.27826488.v6 (2024).
Acknowledgements
This study was funded by the National Key R&D Program of China (grant no. 2022YFF1203004 received by B.H.). This work was also supported by the Beijing Municipal Science and Technology Commission (grant no. Z241100007724005 received by B.H.).
Author information
Authors and Affiliations
Contributions
B.H. conceived the study and supervised the design of all the experiments. B.X. developed HEAD. F.F. and X.M. developed TED. W.Z. provided instructions for artificial intelligence modeling. F.Z. provided instructions on QM calculations. H.Z. provided instructions for the construction of the torsion fragment. H.W., B.Z., Q.X., W.F., X.W., and W.G. supported the evaluation of AI models. Z.L. and Y.W. supported forced field-based optimization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Charlotte Deane and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fan, F., Xi, B., Meng, X. et al. Assessing conformation validity and rationality of deep learning-generated 3D molecules. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69303-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69303-5