Abstract
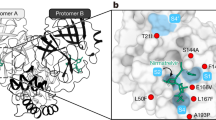
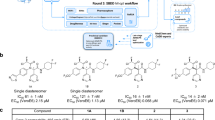
Anecdotal reports about smoking that might prevent SARS-CoV-2 infection inspire the search for nicotine and its pyrolysis products as inhibitors of the SARS-CoV-2 main protease (MPro). This effort leads to the discovery of 3-vinylpyridine as an MPro inhibitor. 3-Vinylpyridine resembles part of nirmatrelvir in binding to MPro but does not involve a critical interaction with residue E166, whose mutation has led to resistance to nirmatrelvir. Integration of the two molecules, followed by a medicinal chemistry campaign, produces several molecules with better in vitro potency than nirmatrelvir. Two lead molecules, YR-C-136 and SR-B-103, display better pharmacokinetic characteristics than nirmatrelvir in virus-challenged male mice and much better antiviral efficacy in virus-challenged female mice. Both molecules maintain high potency in inhibiting the nirmatrelvir-resistant MPro (E166V/L50F) variant. They also exhibit a broad and highly potent antiviral spectrum against most pathogenic coronaviruses. With high in vivo potency, both molecules are potentially standalone pan-antivirals for coronaviruses and may serve as countermeasures for future coronavirus outbreaks.
Similar content being viewed by others
Data availability
All data are available in the main text or the supplementary materials. The crystal structures for MPro complexed with various inhibitors have been deposited into the Protein Data Bank with the following entry codes: 9BS7 (VP), 9BTT (SR-B-51), 9BTF (SR-B-77), 9P6F (SR-B-78), 9BTK (YR-C-108), 9BVW (SR-B-103), 9BVX (YR-C-155), and 9BSR (R-C-136). The source data underlying Figs. 4, 5, and Table 1, Table S2 are provided as a Source Data file. Viral resistance profile data access codes in NCBI SRA: SRR37076225 (wild-type SARS-CoV-2 sequence reads), SRR37076230-SRR37076233 (four biological repeats for SR-B-103 treated viruses), and SRR37076226-SRR37076229 (four biological repeats for YR-C-136 treated viruses). Source data are provided with this paper.
References
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Worobey, M. et al. The emergence of SARS-CoV-2 in Europe and North America. Science 370, 564–570 (2020).
Goel, R. R. et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 374, abm0829 (2021).
Corbett, K. S. et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020).
World Health Organization, COVID-19 Epidemiological Update - 12 March Edition 177, 1–31 (2025).
Mathieu, E. et al. Coronavirus Pandemic (COVID-19). Our World in Data, (2020–2024).
V’Kovski, P., Kratzel, A., Steiner, S., Stalder, H. & Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021).
Kim, D. et al. The Architecture of SARS-CoV-2 Transcriptome. Cell 181, 914–921 e910 (2020).
Morse, J. S., Lalonde, T., Xu, S. & Liu, W. R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 21, 730–738 (2020).
Dai, W. et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335 (2020).
Boby, M. et al. Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors. Science 382, eabo7201 (2023).
Focosi, D., Sullivan, D. J. & Franchini, M. Development of antiviral drugs for COVID-19 in 2025: unmet needs and future challenges. Expert Rev. Anti-Infect. 23, 1–8 (2025).
Yang, K. S., Leeuwon, S. Z., Xu, S. & Liu, W. R. Evolutionary and structural insights about potential SARS-CoV-2 Evasion of Nirmatrelvir. J. Med Chem. 65, 8686–8698 (2022).
Owen, D. R. et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021).
Unoh, Y. et al. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 65, 6499–6512 (2022).
Jiang, X. et al. Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir. Nat. Commun. 14, 6463 (2023).
Zhou, J. et al. Pharmacokinetics and safety of GST-HG171, a novel 3CL protease inhibitor, in Chinese subjects with impaired and normal liver function. Antimicrob. Agents Ch 68, e0053924 (2024).
Zhan, Y. et al. Leritrelvir for the treatment of mild or moderate COVID-19 without co-administered ritonavir: a multicentre randomised, double-blind, placebo-controlled phase 3 trial. Eclinicalmedicine 67, 102359 (2024).
Amani, B. & Amani, B. Efficacy and safety of nirmatrelvir/ritonavir (Paxlovid) for COVID-19: A rapid review and meta-analysis. J. Med. Virol. 95, 1–9 (2023).
Luetkemeyer A. F. et al. Ensitrelvir for the treatment of nonhospitalized adults with COVID-19: Results from the SCORPIO-HR, Phase 3, Randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 80, 1235–1244 (2025).
Chen, X. et al. Effectiveness and Safety of Simnotrelvir/Ritonavir and Nirmatrelvir/Ritonavir in the Treatment of Moderate to Severe COVID-19. Immun. Inflamm. Dis. 13, e70174 (2025).
Lu, H. et al. Efficacy and safety of GST-HG171 in adult patients with mild to moderate COVID-19: a randomised, double-blind, placebo-controlled phase 2/3 trial. Eclinicalmedicine 71, 102582 (2024).
Zhou, Y. et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system. Sci. Adv. 8, eadd7197 (2022).
Nooruzzaman, M. et al. Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections. Nat. Commun. 15, 7999 (2024).
Miyara, M. et al. Low rate of daily active tobacco smoking in patients with symptomatic COVID-19. Qeios 8, 668995 (2020).
Patanavanich, R. & Glantz, S. A. Smoking is Associated with COVID-19 Progression: A Meta-Analysis. Nicotine & Tob. Res., 22, 1653–1656 (2020).
Israel, A., Feldhamer, E., Lahad, A., Levin-Zamir, D. & Lavie, G. Smoking and the risk of COVID-19 in a large observational population study. medRxiv, 2020.06.01.20118877 (2020).
Chattopadhyay, S., Malayil, L., Kaukab, S., Merenstein, Z. & Sapkota, A. R. The predisposition of smokers to COVID-19 infection: A mini-review of global perspectives. Heliyon 9, e17783 (2023).
Rahman, M. et al. Insight into the pulmonary molecular toxicity of heated tobacco products using human bronchial and alveolar mucosa models at air-liquid interface. Sci. Rep. -Uk 12, 16396 (2022).
Yang, K. S. et al. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem 16, 942–948 (2021).
Stevens, N. A. & Borgerding, M. F. GC-AED studies of nicotine fate in a burning cigarette. Anal. Chem. 71, 2179–2185 (1999).
Schechter, I. & Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res Co. 425, 497–502 (2012).
Jin, Z. et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).
Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 368, 409–412 (2020).
Vatansever, E. C. et al. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. USA 118, e2012201118 (2021).
Chen, X. et al. Preclinical evaluation of the SARS-CoV-2 M(pro) inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir. Nat. Microbiol. 9, 1075–1088 (2024).
Lin, C. et al. Structural basis for the inhibition of coronaviral main proteases by ensitrelvir. Structure 31, 1016–1024 e1013 (2023).
Zuckerman, N. S., Bucris, E., Keidar-Friedman, D., Amsalem, M. & Brosh-Nissimov, T. Nirmatrelvir Resistance-de Novo E166V/L50V mutations in an immunocompromised patient treated with prolonged Nirmatrelvir/Ritonavir monotherapy leading to clinical and virological treatment failure-a case report. Clin. Infect. Dis. 78, 352–355 (2024).
Alugubelli, Y. R. et al. A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med Chem. 240, 114596 (2022).
Cao, W. et al. Evaluation of SARS-CoV-2 main protease inhibitors using a novel cell-based assay. ACS Cent. Sci. 8, 192–204 (2022).
Donmez Cakil, Y. et al. Pore-exposed tyrosine residues of P-glycoprotein are important hydrogen-bonding partners for drugs. Mol. Pharm. 85, 420–428 (2014).
Ma, Y. et al. A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 240, 114570 (2022).
Geng, Z. Z. et al. A systematic survey of reversibly covalent dipeptidyl inhibitors of the SARS-CoV-2 main Protease. J. Med Chem. 66, 11040–11055 (2023).
Blankenship, L. R. et al. SARS-CoV-2 Main Protease inhibitors that leverage unique interactions with the solvent exposed S3 site of the enzyme. ACS Med. Chem. Lett. 15, 950–957 (2024).
Khatua, K. et al. Azapeptides with unique covalent warheads as SARS-CoV-2 main protease inhibitors. Antivir. Res. 225, 105874 (2024).
Nagai, K., Fukuno, S., Shiota, M., Tamura, M., Yabumoto, S. & Konishi, H. Differences in transport characteristics and cytotoxicity of Epirubicin and Doxorubicin in HepG2 and A549 Cells. Anticancer Res. 41, 6105–6112 (2021).
Muruato, A. et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. PLoS Biol. 19, e3001284 (2021).
Duan, Y. et al. Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir. Nature 622, 376–382 (2023).
Iketani, S. et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 613, 558–564 (2023).
Lockbaum, G. J. et al. Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188. Viruses 13, 174 (2021).
Song, L. et al. Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors. Acta Pharm. Sin. B 14, 87–109 (2024).
Jacobs, J. et al. Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease. J. Med. Chem. 56, 534–546 (2013).
Xie, X. et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 e843 (2020).
Acknowledgements
We thank Dr. Yohannes Rezenom in the Mass Spectrometry Facility and Prof. Xin Yan of Texas A&M University for helping with the LC-MS characterizations of inhibitors. Funding: National Institutes of Health grant R35GM145351 (W.R.L.). National Institutes of Health grant R21AI164088 (S.X.). National Institutes of Health Grant R21EB032983 (W.R.L.). Welch Foundation Grant A-1715 (W.R.L.). Welch Foundation Grant A-2174 (S.X.). Texas A&M University Advancing Discovery to Market Grant (W.R.L., S.X.). Texas A&M X Grants (W.R.L., S.X.). This work was partially supported by Task Order 75N93024F00002 under Contract 75N93019D00021 from the Respiratory Diseases Branch of the National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
W.R.L., S.X., X.X., and B.W.N. conceived and supervised the research; W.R.L. and S.X. designed all inhibitors; S.A., Y.R.A., V.V., and S.N. synthesized the inhibitors and performed their purification and characterizations; K.K. conducted enzymatic IC50 characterizations of the inhibitors; K.K. and X.S.G. performed in vitro PK analysis of selected inhibitors; D.C. and C.-C.D.C. performed characterizations of cellular IC50 values to engage MPro expressed in HEK293T cells; S.S. and C.-C.D.C. expressed and purified MPro (E166V/L50F), SARS-CoV MPro, and MERS-CoV MPro; L.R.B., K.Y., and D.R. conducted the X-ray protein crystallography analysis; S.K. characterized antiviral potency of inhibitors in cell culture; H.X. and B.K.K. conducted in vivo antiviral studies and mouse tissue analysis, D.H.W. performed the histopathologic analysis and scoring; B.L.H. conducted the in vitro antiviral potency test on SARS-CoV-2 USA_WA1/2020, SARS-CoV-2 Delta B.1.617.2 AY.4, SARS-CoV-2 Omicron BQ.1, HCoV-Alpha 229E, HCoV-Beta OC43, SARS Urbani, and MERS-CoV EMC variants. All authors participated in data analysis. W.R.L., S.X., X.X., and B.W.N. drafted the manuscript with the assistance of K.K., D.C., S.A., Y.R.A., V.V., and L.R.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jun Wang and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khatua, K., Atla, S., Coleman, D. et al. From nicotine to SARS-CoV-2 antivirals with potent in vivo efficacy and a broad anti-coronavirus spectrum. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69527-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69527-5