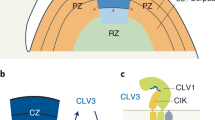
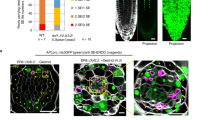
Abstract
Secreted CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptide ligands dimension the stem cell niche of Arabidopsis shoot meristems by signalling through redundant and cross-compensating CLAVATA1 (CLV1)-type receptor kinases. In the root meristem, the CLV1 homologues BARELY ANY MERISTEM 1 (BAM1) and BAM2 drive CLE13/16-mediated formative divisions that produce the ground tissue layers. Here we report that BAM1/2 are also required to initiate the vascular phloem lineage and that cross-compensation between CLV1-type receptors as observed in the shoot does not operate similarly in the root. Rather, we find that BAM3-mediated CLE45 signalling antagonizes BAM1/2-mediated CLE11/12/13 signalling in the phloem initials but not in the ground tissue. We further observe spatiotemporally contrasting CLE signalling requirements for phloem initiation and differentiation, which are shaped by the SHORT ROOT (SHR) pathway. Our findings thus suggest an intricate quantitative interplay between distinct and antagonistic CLE signalling pathways that organizes tissue layer formation in the Arabidopsis root meristem.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions are presented in the main figures and the Extended Data. Materials are available upon request. The RNA sequencing reads are deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under submission ID SUB14584842, sample IDs SAMN42396145–SAMN42396186. Source data are provided with this paper.
References
Kitagawa, M. & Jackson, D. Control of meristem size. Annu. Rev. Plant Biol. 70, 269–291 (2019).
Willoughby, A. C. & Nimchuk, Z. L. WOX going on: CLE peptides in plant development. Curr. Opin. Plant Biol. 63, 102056 (2021).
Nimchuk, Z. L. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS Genet. 13, e1006681 (2017).
Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A. & Meyerowitz, E. M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049 (2015).
DeYoung, B. J. et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16 (2006).
Schlegel, J. et al. Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. Elife 10, e70934 (2021).
Hord, C. L., Chen, C., Deyoung, B. J., Clark, S. E. & Ma, H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18, 1667–1680 (2006).
Crook, A. D. et al. BAM1/2 receptor kinase signaling drives CLE peptide-mediated formative cell divisions in Arabidopsis roots. Proc. Natl Acad. Sci. USA 117, 32750–32756 (2020).
Hazak, O. et al. Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep. 18, 1367–1381 (2017).
Qian, P. et al. The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 4, 1071–1081 (2018).
Qian, P. et al. A Dof-CLE circuit controls phloem organization. Nat. Plants 8, 817–827 (2022).
Roman, A. O. et al. HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides. Nat. Commun. 13, 876 (2022).
Zhang, H., Lin, X., Han, Z., Qu, L. J. & Chai, J. Crystal structure of PXY–TDIF complex reveals a conserved recognition mechanism among CLE peptide–receptor pairs. Cell Res. 26, 543–555 (2016).
Carbonnel, S., Cornelis, S. & Hazak, O. The CLE33 peptide represses phloem differentiation via autocrine and paracrine signaling in Arabidopsis. Commun. Biol. 6, 588 (2023).
Jun, J. et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736 (2010).
Kinoshita, A. et al. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825 (2007).
Depuydt, S. et al. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl Acad. Sci. USA 110, 7074–7079 (2013).
Anne, P. & Hardtke, C. S. Phloem function and development—biophysics meets genetics. Curr. Opin. Plant Biol. 43, 22–28 (2017).
Bonke, M., Thitamadee, S., Mahonen, A. P., Hauser, M. T. & Helariutta, Y. APL regulates vascular tissue identity in Arabidopsis. Nature 426, 181–186 (2003).
Rodriguez-Villalon, A. et al. Molecular genetic framework for protophloem formation. Proc. Natl Acad. Sci. USA 111, 11551–11556 (2014).
Czyzewicz, N. et al. Antagonistic peptide technology for functional dissection of CLE peptides revisited. J. Exp. Bot. 66, 5367–5374 (2015).
Denyer, T. et al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48, 840–852 e5 (2019).
Jean-Baptiste, K. et al. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell 31, 993–1011 (2019).
Ryu, K. H., Huang, L., Kang, H. M. & Schiefelbein, J. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 179, 1444–1456 (2019).
Shahan, R. et al. A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Dev. Cell 57, 543–560.e9 (2022).
Shulse, C. N. et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 27, 2241–2247.e4 (2019).
Zhang, T. Q., Xu, Z. G., Shang, G. D. & Wang, J. W. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant 12, 648–660 (2019).
Stahl, Y. et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23, 362–371 (2013).
Rodriguez-Villalon, A., Gujas, B., van Wijk, R., Munnik, T. & Hardtke, C. S. Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142, 1437–1446 (2015).
Wang, Q. et al. A phosphoinositide hub connects CLE peptide signaling and polar auxin efflux regulation. Nat. Commun. 14, 423 (2023).
Diaz-Ardila, H. N., Gujas, B., Wang, Q., Moret, B. & Hardtke, C. S. pH-dependent CLE peptide perception permits phloem differentiation in Arabidopsis roots. Curr. Biol. 33, 597–605.e3 (2023).
Gujas, B. et al. A reservoir of pluripotent phloem cells safeguards the linear developmental trajectory of protophloem sieve elements. Curr. Biol. 30, 755–766.e4 (2020).
Hardtke, C. S. Phloem development. New Phytol. 239, 852–867 (2023).
Ren, S. C. et al. CLE25 peptide regulates phloem initiation in Arabidopsis through a CLERK-CLV2 receptor complex. J. Integr. Plant Biol. 61, 1043–1061 (2019).
Fukuda, H. & Hardtke, C. S. Peptide signaling pathways in vascular differentiation. Plant Physiol. 182, 1636–1644 (2020).
Fukuda, H. & Ohashi-Ito, K. Vascular tissue development in plants. Curr. Top. Dev. Biol. 131, 141–160 (2019).
Breda, A. S. et al. A cellular insulator against CLE45 peptide signaling. Curr. Biol. 29, 2501–2508.e3 (2019).
Fan, P. et al. The receptor-like kinases BAM1 and BAM2 are required for root xylem patterning. Proc. Natl Acad. Sci. USA 118, e2022547118 (2021).
Yang, S. et al. A novel chemical inhibitor of polar auxin transport promotes shoot regeneration by local enhancement of HD–ZIP III transcription. New Phytol. 235, 1111–1128 (2022).
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010).
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000).
Nakajima, K., Sena, G., Nawy, T. & Benfey, P. N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001).
Levesque, M. P. et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4, e143 (2006).
Sebastian, J. et al. PHABULOSA controls the quiescent center-independent root meristem activities in Arabidopsis thaliana. PLoS Genet. 11, e1004973 (2015).
Kang, Y. H. & Hardtke, C. S. Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling. EMBO Rep. 17, 1145–1154 (2016).
Hu, C. et al. A CLE–BAM–CIK signalling module controls root protophloem differentiation in Arabidopsis. New Phytol. 233, 282–296 (2022).
Nodine, M. D., Yadegari, R. & Tax, F. E. RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12, 943–956 (2007).
Moret, B., Marhava, P., Aliaga Fandino, A. C., Hardtke, C. S. & Ten Tusscher, K. H. W. Local auxin competition explains fragmented differentiation patterns. Nat. Commun. 11, 2965 (2020).
Kang, Y. H., Breda, A. & Hardtke, C. S. Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 144, 272–280 (2017).
Kim, H. et al. SHORTROOT-mediated intercellular signals coordinate phloem development in Arabidopsis roots. Plant Cell 32, 1519–1535 (2020).
Aida, M. et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Galinha, C. et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007).
Mahonen, A. P. et al. PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129 (2014).
Kinoshita, A. et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920 (2010).
Mizuno, S. et al. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50, 751–766 (2007).
Shimizu, N. et al. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 208, 1104–1113 (2015).
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A. & Simon, R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176 (2010).
Jones, D. S., John, A., VanDerMolen, K. R. & Nimchuk, Z. L. CLAVATA signaling ensures reproductive development in plants across thermal environments. Curr. Biol. 31, 220–227.e5 (2021).
Graeff, M. et al. A single-cell morpho-transcriptomic map of brassinosteroid action in the Arabidopsis root. Mol. Plant 14, 1985–1999 (2021).
Marhava, P. et al. Plasma membrane domain patterning and self-reinforcing polarity in Arabidopsis. Dev. Cell 52, 223–235.e5 (2020).
Acknowledgements
We thank Z. Nimchuk and X. Gou for sharing genetic materials; Z. Nimchuk for comments on the manuscript; and P. Cattaneo for technical assistance. This study was supported by Swiss National Science Foundation grant 310030_207876 awarded to C.S.H. and a CSC scholarship awarded to H.Z.
Author information
Authors and Affiliations
Contributions
H.Z. and C.S.H. designed the project and drafted the manuscript. H.Z., Q.W. and N.B.-T. performed experiments and analysed data. All authors contributed to the assembly and the revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Jungmook Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 cle45/bam3 second-site mutations restore phloem formation in bam12 double mutants.
a, Root length measurements for 7-day-old seedlings of the indicated genotypes. Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (lower case letters) were determined by ordinary one-way ANOVA followed by Tukey’s test, two-sided, p < 0.0001. n = 8–15 independent biological replicates. b, Representative histological cross sections (toluidine blue-stained) from indicated genotypes. Orange arrowheads point out protophloem sieve element cell files.
Extended Data Fig. 2 Expression patterns of genes investigated in this study.
Schematic representation of root tip expression patterns of the indicated genes, obtained from aggregation of multiple independent single-cell mRNA sequencing experiments of Arabidopsis Col-0 wildtype roots (https://rootcellatlas.org). Note the differences in expression level scales.
Extended Data Fig. 3 CLE45 treatments do not affect the SHR pathway.
a, Confocal microscopy images of Col-0 wildtype root meristems grown on 20 nM CLE45 peptide in the absence or presence of ZIC2 (25 mM). b, Confocal microscopy images of Col-0 root meristems expressing the CVP2::NLS-VENUS protophloem sieve element marker, grown on ZIC2 (25 mM) as compared to control. c, Confocal microscopy images of Col-0 root meristems (propidium iodide staining, red fluorescence) expressing various markers of the SHR pathway (green fluorescence), grown on 15 nM CLE45 peptide as compared to controls. Seedlings were 5-day-old throughout.
Extended Data Fig. 4 phb/phv second-site mutations partly rescue the brx mutant phenotype.
a, Confocal microscopy images of 5-day-old root meristems of the indicated genotypes, showing optical longitudinal sections (top) and cross sections (bottom). Orange arrowheads in longitudinal sections point out protophloem sieve element cell files, yellow arrowheads indicate protophloem sieve element precursors that fail to differentiate. Colored dots in cross sections indicate cell files derived from the phloem-procambium precursor stem cell (see Fig. 1a). b, Root length measurements for 7-day-old seedlings of the indicated genotypes. Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (lower case letters) were determined by ordinary one-way ANOVA followed by Tukey’s test, two-sided, p ≤ 0.0038. n = 15–79 independent biological replicates. c, Images of representative 7-day-old seedlings of the indicated genotypes.
Extended Data Fig. 5 bam3 brx double mutants are hypersensitive to CLE peptides.
a-c, Root length measurements for 7-day-old seedlings of the indicated genotypes, grown in the presence of a set of CLE peptides (all 20 nM). Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (asterisks) compared to untreated control were determined by ordinary one-way ANOVA followed by Tukey’s test, two-sided, *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001. n = 5–20 independent biological replicates. d, Root length measurements for 7-day-old seedlings of the indicated genotypes. Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (lower case letters) were determined by ordinary one-way ANOVA, p < 0.0001. n = 18–35 independent biological replicates.
Extended Data Fig. 6 Reducing CLE45-BAM3 signalling in initials permits initiation of the phloem lineage.
a-b, Confocal microscopy images of a root meristem from a bam3 brx seedling expressing a SHR::BAM3-CITRINE transgene (b) as compared to the mutant control (a). c, Confocal microscopy showing the transgenic BAM3-CITRINE signal (yellow fluorescence) by itself (top) and overlayed with calcofluor cell wall staining (bottom). d, Confocal microscopy images of a bam1 bam3 brx root meristem. e-f, Similar to b-c, for a SCR::BAM3-CITRINE transgene. g-h, Confocal microscopy images of plt1 plt2 root meristems grown in the presence of ZIC2 (h) (25 mM) as compared to untreated control (g). i, Confocal microscopy images of a bam3 plt1 plt2 root meristem. j, Confocal microscopy images of plt1 plt2 and bam3 plt1 plt2 root meristems grown in the presence of 20 nM CLE45 peptide. k-l, Root length measurements for 7-day-old seedlings of the indicated genotypes, grown in the presence of 20 nM CLE45 peptide as compared to untreated controls. Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (asterisks or lower-case letters) compared to untreated control (k) or wildtype (l) were determined by ordinary one-way ANOVA followed by Tukey’s test, two-sided, p < 0.0001. n = 10–45 independent biological replicates. m, Confocal microscopy images of Col-0 wildtype root meristems grown in the presence of 20 nM CLE45 peptide applied from germination onwards or at later timepoints. Confocal images were obtained from 5-day-old seedlings. Orange and grey arrowheads in longitudinal sections point out protophloem sieve element cell files and the xylem axis, respectively, yellow arrowheads indicate protophloem sieve element precursors that fail to differentiate. Colored dots in cross sections indicate cell files derived from the phloem-procambium precursor stem cell (see Fig. 1a).
Extended Data Fig. 7 RPK2 cannot replace BAM3 as a CLE peptide receptor.
a, Amino acid sequence alignment of the prototypical CLV1-type CLE peptide receptors and the proposed alternative receptor, RPK2. Green arrowheads point out amino acid residues that have been implicated in CLE peptide interaction. b, Confocal microscopy images of 5-day-old bam3 and bam3 brx mutant root meristems (propidium iodide staining, magenta fluorescence) expressing an RPK2-CITRINE fusion protein (green fluorescence) in the phloem pole, under control of the BAM3 promoter. c, Root length measurements for 8-day-old seedlings of the indicated genotypes, grown in the presence of 20 nM CLE45 or CLE33 peptide. Box plots display 2nd and 3rd quartiles and the median, whiskers indicate maximum and minimum. Statistically significant differences (asterisks) compared to untreated control were determined by ordinary one-way ANOVA followed by Tukey’s test, two-sided, ***: p < 0.002; ****: p < 0.0001. n = 28–61 independent biological replicates.
Supplementary information
Supplementary Table 1
Gene expression levels in RNAseq.
Supplementary Table 2
Differential expression analysis of RNAseq.
Supplementary Table 3
Gene ontology (GO) analysis of RNAseq.
Supplementary Table 4
List of mutant alleles.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, H., Wang, Q., Blanco-Touriñán, N. et al. Antagonistic CLE peptide pathways shape root meristem tissue patterning. Nat. Plants 10, 1900–1908 (2024). https://doi.org/10.1038/s41477-024-01838-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-024-01838-1
This article is cited by
-
Peptide hormones in plants
Molecular Horticulture (2025)