Abstract
Septoria tritici blotch (STB), caused by the Dothideomycete fungus Zymoseptoria tritici, is one of the most damaging diseases of bread wheat (Triticum aestivum)1 and the target of costly fungicide applications2. In line with the fungus’s apoplastic lifestyle, STB resistance genes isolated to date encode receptor-like kinases (RLKs) including a wall-associated kinase (Stb6) and a cysteine-rich kinase (Stb16q)3,4. Here we used genome-wide association studies on a diverse panel of 300 whole-genome shotgun-sequenced wheat landraces (WatSeq consortium5) to identify a 99-kb region containing six candidates for the Stb15 resistance gene. Mutagenesis and transgenesis confirmed a gene encoding an intronless G-type lectin RLK as Stb15. The characterization of Stb15 exemplifies the unexpected diversity of RLKs conferring Z. tritici resistance in wheat.
Similar content being viewed by others
Main
The domestication of wheat 10,000 years ago heralded the dawn of modern agriculture in western Eurasia while providing an opportunity for the specialization of an uninvited guest: the fungal pathogen Zymoseptoria tritici6. Understanding and bolstering genetic resistance to this pathogen could aid in reclaiming ~24 million tonnes of wheat yield lost to Septoria tritici blotch (STB) each year1,7.
During its interaction with wheat, Z. tritici colonizes the apoplast through the stomata and commences a period of asymptomatic growth wherein effectors are released: molecules that suppress host defences or make the host amenable to colonization8. Host resistance proteins may directly or indirectly recognize these effectors and modulate defence responses, described in apoplastic interactions as effector-triggered defence or the “invasion model”9,10,11. If undetected, the pathogen switches to its necrotrophic life stage, resulting in the release of host nutrients and the rapid growth and proliferation of the pathogen12. Symptoms ultimately manifest as necrotic lesions on the leaf surface containing pycnidia (asexual fruiting bodies), which produce conidia that may disperse up to one metre by rain splash, allowing further cycles of colonization and thus quick progress of the disease13.
Twenty-three major genes controlling isolate-specific resistance to STB (Stb genes) have been mapped in wheat14,15,16, but Stb gene cloning has lagged behind efforts for other wheat diseases. Stb6 on chromosome 3AS, conferring race-specific resistance to Z. tritici3,17, encodes a wall-associated receptor kinase, a subfamily within the receptor-like kinase (RLK) family in plants, with a galacturonan-binding domain3. Stb16q on chromosome 3D18 encodes a cysteine-rich receptor kinase with two DUF26 domains4. Stb15 is a major gene for resistance to Z. tritici isolate IPO88004, mapped to a 36 cM region in the cultivar Arina19. Stb15 is a good candidate for cloning and thus for studying the biology of STB resistance owing to its large phenotypic effect resulting in full resistance to avirulent isolates, which is rare among Stb genes14, and because it is widely present throughout European wheat cultivars (although it no longer confers field resistance in the United Kingdom20 and possibly elsewhere).
Here we apply genome-wide association studies (GWAS) to map resistance to STB in the diverse Watkins collection of pre-Green Revolution wheat landraces, which provides a unique opportunity to study interactions with Z. tritici in a well-adapted yet highly genetically diverse context21,22,23. This panel represents the genetic and phenotypic variability of wheat before the Green Revolution, primarily throughout its pre-Columbian range across Eurasia and North Africa. Whole-genome shotgun sequencing of a core collection of 300 Watkins lines, selected to maximize genetic representation, offers access to the entire genetic diversity of the panel and permits mapping of resistance genes at major quantitative trait loci for use in research and breeding5.
STB symptoms elicited by the Z. tritici isolates IPO323, avirulent to Stb6 (ref. 17), and IPO88004, avirulent to Stb15 (ref. 19), were scored across 300 Watkins landraces (Fig. 1a). This core panel was selected to maximize genetic representation24. Leaf damage (necrosis and chlorosis) and pycnidial coverage are usually, but not always, correlated25. Both phenotypes were recorded at five or six time points for calculation of the area under the disease progress curve (AUDPC), followed by logit transformation and linear mixed modelling (Supplementary Tables 1–4).
a, Quantitative variation in pycnidia and necrosis phenotypes. Pictured are leaves arranged by pycnidial coverage (0–100%). b, Effects of Stb6 and Stb15 on pycnidia scores with Z. tritici isolates IPO88004 and IPO323. The logit pycnidial area under the disease progress curve (pAUDPC) values of the axes are back-transformed to give pAUDPC (0–100%). c, Manhattan plots showing the association of logit pAUDPC in response to Z. tritici isolates IPO323 (left) and IPO88004 (right) with SNPs mapped to Chinese Spring, in terms of Wald test P values, not adjusted for multiple comparisons. LD blocks associated with STB resistance are drawn as arrows beneath the chromosomes (marked in bold) with the 6A Stb15 candidate gene marked in orange and Stb6 in purple. The large interval for the locus STBWat1 is also shown.
A single nucleotide polymorphism (SNP) matrix generated from whole-genome shotgun sequencing data of wheat cultivars and landraces was mapped to Chinese Spring and employed for GWAS5. As a positive control to ensure the suitability of the experimental system for mapping via this method, Stb6 was successfully restricted to a discrete genomic interval in the core Watkins panel. We screened IPO323 on the Watkins core panel and ten control cultivars (Supplementary Table 5), including Chinese Spring, which has the functional allele of Stb6 (ref. 3) (Fig. 1b). An interval on chromosome 3A was associated with both leaf damage and pycnidia phenotypes (Fig. 1c (pycnidia) and Supplementary Fig. 1 (leaf damage)). SNPs in the 3A locus were highly associated with pycnidia, with a −log10 P value of almost 30. Within this region, a linkage disequilibrium (LD) block extending from 26.10 to 27.50 Mb was identified. A smaller haploblock within it was most highly associated with resistance, from 26,035,170 to 26,238,727 bp (Supplementary Fig. 2). This 203.6-kb region contained six genes, including Stb6 (Fig. 1c).
We then proceeded to identify Stb15 by inoculating the panel with IPO88004 and employing GWAS. Three regions were associated with STB phenotypes (Fig. 1c (pycnidia) and Supplementary Fig. 3 (leaf damage)). A locus on 6AS had the highest P value for both pycnidia and damage traits and spanned a 99.1-kb region between 48,550,326 and 48,599,421 bp containing six genes in Chinese Spring. Gene sequences were compared between ArinaLrFor, which has the Stb15 resistance phenotype, and Chinese Spring, which does not; both lines have high-quality, annotated genome sequences. This comparison, combined with the correlation of haplotypes with the responses of landraces to IPO88004, excluded five of these genes (Supplementary Table 6 and Supplementary Fig. 4). The remaining gene, TraesCS6A02G078700 (Chinese Spring)/TraesARI6A03G03215890 (ArinaLrFor), is predicted to encode an RLK and is strongly associated with isolate-specific resistance to IPO88004 but not with Stb6 resistance to IPO323 (Fig. 1b), so it was selected as the most likely candidate for Stb15.
We also observed a significant association of pycnidia cover of IPO88004 with a locus on chromosome 2B from position 755 to 775 Mb containing 241 genes. When we removed the masking effect of lines carrying Stb15, the significance of the 2BL resistance increased 1,000-fold (Supplementary Fig. 5). Stb9 has previously been mapped to 2BL26 but is outside of this locus (at ~808 Mb)27, and accessions that display resistance to IPO89011, an isolate that is avirulent on Stb9, are not all resistant to IPO88004 (refs. 20,28) (Supplementary Table 7). The LD block therefore appears to be a new locus for resistance to Z. tritici, temporarily designated as STBWat1.
TraesARI6A03G03215890 in ArinaLrFor was confirmed as Stb15 by a combination of mutagenesis and transgenesis and was shown to be a lectin receptor kinase (LecRK). We screened 3,308 plants from 307 M2 families of an EMS-derived mutant population of cv. Arina, the donor of Stb15 in ArinaLrFor, for resistance to IPO88004 and identified three independent susceptible mutants (Fig. 2a,b). All three of these mutant plants had one non-synonymous transition mutation in the open reading frame of the Stb15 candidate. The gene encodes a G-type LecRK29 with an intracellular serine/threonine receptor-like protein kinase and three extracellular domains: a mannose-specific bulb-type lectin (BTL), an S-locus glycoprotein (SLG) and a plasminogen/apple/nematode (PAN) domain. All three of the induced mutations resulted in replacement by larger amino acids in the BTL and kinase domains. In an AlphaFold model (Fig. 2c and Supplementary Fig. 6; predicted local distance difference test (pLDDT) score, 80.79), all three residues were in locations where mutations would be predicted to cause disruption to the protein structure. To confirm the function of the candidate gene, we synthesized a 10.9-kb genomic sequence containing 2 kb and 1.5 kb of 5′ and 3′ regulatory sequence from Arina, inserted this sequence into a binary vector and transformed wheat cv. Fielder, which is susceptible to isolate IPO88004. We obtained two independent homozygous single-copy T2 transgenic lines that conferred resistance to pycnidium formation by the Stb15-avirulent isolate IPO88004 but not IPO92006 (Stb15-virulent), whereas their respective nulls were susceptible to both isolates, indicating that the isolated gene sequence is sufficient to confer the Stb15 phenotype. Transgenic lines with four copies or six to eight copies of Stb15 were also resistant to IPO88004 relative to the controls (Fig. 2d, Supplementary Fig. 7 and Supplementary Tables 8 and 10–12). There was no evidence that Stb15 copy number affected the resistance phenotype (Supplementary Table 10). By contrast, variation in damage was neither statistically significant nor isolate-specific (Supplementary Fig. 7c,f and Supplementary Table 9).
a, The functional resistance allele of Stb15 in wheat cv. Arina and ArinaLrFor compared to the susceptible allele in cv. Chinese Spring. The predicted exons and introns are shown as rounded rectangles and lines, respectively, for Chinese Spring (RefSeq v.1.1 (ref. 84)) and ArinaLrFor (Methods). Domains are highlighted: SP, signal peptide; TM, transmembrane; S/TPK, serine/threonine receptor-like protein kinase. InterProScan was used to predict protein domains, with additional adjustments made for ArinaLrFor on the basis of the AlphaFold model (c). The white boxes indicate untranslated regions. The sequence variants of three EMS-induced loss-of-function mutants inoculated with Z. tritici isolate IPO88004 are indicated. b, STB phenotypes of the three EMS mutants. c, AlphaFold-augmented 3D structural model of Stb15. The domains are coloured as in a. The locations of the three EMS-induced mutations are shown in red and indicated by labelled red arrows. d, Cultivar Fielder stably transformed with an Stb15 construct and inoculated with isolate IPO88004. Null-2.1:0 is a null wherein the transgene segregated out in the T2 family, while GRF-5:4 was transformed with the same vector backbone minus Stb15. Further details about the transgenic line names are provided in Supplementary Table 10. The copy number of Stb15 is given as a fixed number or range. In b and d, the leaf sections outlined with dashed boxes and labelled 1, 2 and 3 are enlarged on the right side for improved visibility of pycnidia.
There are two alleles of Stb15 in the wheat pangenome and a further two non-functional alleles in Watkins landraces (Supplementary Table 4). Alleles were defined on the basis of SNP distance (Supplementary Fig. 4 and Supplementary Tables 13 and 14). The functional ArinaLrFor allele of Stb15 is present across the geographic (Fig. 3a) and genetic (Fig. 3b) diversity of the Watkins core 300 collection, although it occurs in only 15% of landraces. It is often present alongside Stb6, which is more common (78%). A total of 14% of landraces displayed resistance to Z. tritici, which could not be explained by either gene. Unexplained resistance to IPO88004 (36 landraces) could be due to STBWat1 (Fig. 1c). Stb15 is also present in 35% of European cultivars tested using KASP markers (Supplementary Table 15).
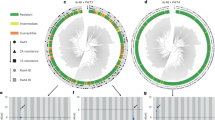
a, Distribution of Stb6 and Stb15 in the Watkins 300 core collection. The map indicates the coordinates of local markets from which grain of landraces was obtained. Only countries from which landraces were collected are labelled. The country abbreviations are expanded in Supplementary Text 1. b, Principal component analysis plot of Axiom array SNPs from 300 Watkins landraces with lines containing predicted functional alleles of Stb6 (purple), Stb15 (orange), both (pink) or neither (grey) indicated. c, Maximum likelihood phylogenetic tree of proteins with homology to the ArinaLrFor (ArinaLF) Stb15-encoded allele from selected Poaceae species, including the wheat reference genome Chinese Spring (CS). The smallest non-repetitive (‘inner’) clade containing Stb15 is shown. The intron/exon structure of Stb15 homologues and their relative nucleotide lengths are presented (arrows indicate exon coding sequences; lines indicate introns). Species names and chromosomes are given; Un indicates homologues within scaffolds that have not yet been mapped to chromosomes. Gene IDs for homologues are given in Supplementary Table 17. d, Protein alignment of the homologues from c with alignment gaps, sequence conservation and predicted protein domains indicated. Taller, greener bars in the conservation panel indicate more conserved regions.
Forty-eight proteins from 16 Poaceae species shared homology with the Arina/ArinaLrFor Stb15 protein (Supplementary Fig. 8 and Supplementary Tables 16 and 17). Homologous genes encoding the protein sequences were found to be conserved across the Group 6 chromosomes within the Triticeae (Fig. 3c) but were also present on other chromosomes, especially Groups 3, 4 and 7 (Supplementary Fig. 8). Microsynteny of the Stb15 mapping interval was observed in wheat lines and relatives (Supplementary Fig. 9).
We detected intron/exon structural diversity in gene annotations of proteins with homology to Stb15 across the Poaceae, including within Triticum aestivum (Fig. 3c). The functional allele of Stb15 is intronless, while in cv. Chinese Spring, lacking the Stb15 phenotype, the gene has four introns. An intronless gene structure was also observed in gene annotations of 22 homologous proteins both within the Triticeae tribe (Triticum, Aegilops and Hordeum) and beyond it (Brachypodium and Avena), especially on Group 3 and 6D/6S chromosomes. Stb15 clustered most closely to the Chinese Spring 6D and Aegilops bicornis 6S homologues, suggesting that the functional allele of Stb15 may have originated from the D or S genomes, which share high sequence homology30.
Within the inner Stb15 clade, the kinase and PAN domains were highly conserved, while the region spanning the BTL and SLG domains was variable (Fig. 3d). This suggests that this region may be under diversifying selection.
Characterization of the third Stb gene from a distinct subclass of the RLK protein family has the potential to enhance molecular understanding of the wheat–Z. tritici interaction, providing new opportunities for research and disease control. In addition, this research demonstrates the power of GWAS to greatly accelerate gene cloning for traits that are poorly understood at the molecular level. One of the factors that may limit the success of GWAS is population structure31. In this study, the presence of Stb15 and Stb6 in Watkins landraces that spanned the breadth of the genetic diversity of the panel was probably decisive to their successful mapping (Fig. 3a,b). Such wide distributions of allelic variation across the full range of relevant germplasm allow the effects of genes of interest to be separated from those of kinship. Such a distribution may be more likely for genes that were introduced early into cultivated hexaploid wheat, which appears to be the case for Stb6, known from both Europe and East Asia32. Likewise, Watkins lines with Stb15 were obtained across the breadth of Eurasia as well as North Africa.
The diversity of intron/exon gene structures among Stb15 homologues is unusual when compared with nucleotide-binding leucine-rich repeats, for which gene structures tend to be conserved33. Leucine-rich-repeat membrane-anchored proteins without intracellular kinases control resistance to the related Dothidiomycete fungus Cladosporium fulvum, the causal agent of leaf mould in tomato34, many of which35 share the intronless open reading frame exhibited by the functional allele of Stb15. Possibly, intronless gene structures have been conserved while intron gain has occurred in, for example, the Chinese Spring allele.
Stb15 has more extracellular domains than Stb6 or Stb16q, and the diversity of RLK subclasses conferring resistance is unusual compared with Cf genes but similar to genes conferring resistance to blackleg disease in Brassica species36,37. Equally, there are similarities shared by Stb proteins: they are transmembrane proteins with extracellular domains with a putative sugar-binding function and an intracellular kinase. Both the DUF26 domains of Stb16q and the G-type lectin domain of Stb15 probably bind mannose, a building block of mannan found in cell walls of both fungi38 and wheat39. The detection of a conserved pathogen-associated molecular pattern (PAMP) fits the function of lectins, which form part of basal plant immunity and are involved in stomatal innate immunity responses in Arabidopsis thaliana40, and LecRKs confer non-host or marginal host resistance to leaf rust in barley41. This role could make Stb15 a target for suppression by the pathogen, or it could be part of a guard/guardee pair42, triggering isolate-specific resistance. Alternatively, the interaction could resemble that of the tomato receptor Cf-4 and Avr4, a passive C. fulvum effector that binds chitin to avoid breakdown by the host plant34. Another possibility is that Stb15 binds glycoproteins; AFP1 in maize was previously thought to bind chitin but in fact interacts with chitin deacetylases, most likely via their mannosylated group43.
LecRKs have been found to bind to secreted proteins (for example, a Phytophthora spp. effector44), which may also be the case for Stb15. A candidate gene for AvrStb15 encoding a small secreted protein has been suggested45, but further work will be needed to determine the nature of its interaction with Stb15. There is thus far no evidence of a direct interaction between Stb6 and AvrStb6, also encoding a cysteine-rich small secreted protein3,46,47. In conclusion, our study highlights the importance of elucidating the diverse roles of Stb genes in defence induction for understanding the genetic basis of resistance in this economically important pathosystem.
Methods
Plant and pathogen material
Of 826 lines in the Watkins collection, we used a core set of 300 lines representing the majority of genetic variation in spring growth types24. Control lines were included in all assays, with lines selected for known responses to STB or economic importance (Supplementary Table 5). Both Arina and ArinaLrFor were used for analyses or experiments on Stb15 as they carry the same allele. ArinaLrFor was derived from an Arina × Forno cross and further backcrossing with Arina48, where cv. Forno is susceptible to IPO88004, so the resistance in ArinaLrFor should come from the Arina allele of Stb15. Seeds of ArinaLrFor (PANG0001) are available from the Germplasm Resources Unit, John Innes Centre (JIC) (https://www.jic.ac.uk/research-impact/germplasm-resource-unit/). The Z. tritici isolates IPO323 (virulent on Stb15) and IPO88004 (virulent on Stb6) were used for their avirulence to Stb6 (refs. 3,17) and Stb15 (ref. 19), respectively. IPO323 was isolated in the Netherlands in 198149 and IPO88004 in Ethiopia in 198850. A third isolate, IPO90012 from Mexico51, was also included as a control virulent on both Stb6 and Stb15; it is also avirulent on Stb11 (ref. 14).
Design and infection protocol for pathology assays at the JIC
An alpha lattice design52 was used for pathology experiments on 300 WatSeq landraces with five replicates per line across incomplete blocks (40-well seedling trays) in a large controlled environment room (CER) trial. This allowed the effects of tray and position in the CER to be estimated. The design was generated using Gendex (http://designcomputing.net/gendex/).
Standard pathology methods were used for the inoculation of wheat seedlings (ref. 53, following ref. 50). Seeds of the lines tested were pre-germinated in Petri dishes on 90 mm filter paper (Whatman International) containing 4 ml of 0.2 ppm gibberellic acid. The Petri dishes were placed in the dark at room temperature for 48 hours, then moved to the lab bench in daylight for a further 24 hours. Germinated seeds were planted in John Innes peat-based F2 compost in 40-well trays. The trays were placed in a Conviron CER with a 16-hour photoperiod: day temperature, 18 °C; night temperature, 12 °C; photosynthetic photon flux density, 350 µE m−2 at plant height. When the second leaf was fully expanded (Zadoks Growth Stage 12), usually around 14 days after germination, inoculum was prepared.
Sporulating cultures of Z. tritici were grown on potato dextrose agar plates for five to seven days under near-ultraviolet light (Snijders Micro Clima-Series Economic Lux Chamber) for 16 h per day at 18 °C. The cultures were flooded with 3 ml of sterile distilled water and scraped to release conidia. The concentration of conidial suspension was adjusted to the desired value of 106 spores per ml. Conidial concentration was assessed using a Fuchs-Rosenthal counting chamber (Hawksley). Two drops of polyoxyethylene-sorbitan monolaurate (Tween-20; Sigma-Aldrich Chemie) were added per 50 ml of spore suspension.
Later-formed leaves were cut away so that only the primary seedling leaf remained. The seedlings were evenly sprayed with the spore suspension (20 ml per tray) on a turntable (made at JIC), using a Wiz Mini Air Compressor spray gun kit (Clarke Tools).
The trays were placed on matting within propagators, with two trays per propagator. This allowed the trays to be watered from underneath to prevent inoculum from washing off. The propagators were closed and covered with black plastic for dark incubation. The black plastic was removed after 48 hours, and the propagator lids were kept over the trays until seven days after inoculation to increase humidity and therefore the success of Z. tritici infection. New leaf growth was cut back twice per week to keep inoculated leaves alive and facilitate scoring.
The percentage of leaf area covered by pycnidia and leaf damage was scored by eye four to six times at intervals of two to five days over a period of 10 to 32 days post inoculation, depending on disease progress. Damage was defined as the combined area of necrosis and chlorosis. For imaging, leaves were mounted on A4 paper and scanned with a Canon LiDE 120 scanner at 600 DPI using the Canon IJ Scan Utility2 software. The standardized A4 image size allowed the dimensions of cropped images to be calculated using Adobe Illustrator.
Statistical analysis of STB phenotypes
The AUDPC was calculated for each leaf by summing the areas of trapezia between pairs of scoring days on a graph of disease severity over time. The AUDPC data were analysed for the effects of line, isolate and experimental design factors using linear mixed modelling with random and fixed effects, via the package lmerTest54 in R v.4.2.2. If only fixed effects were involved, the native R analysis-of-variance function aov was used. F-tests were conducted to determine concise fixed models that explained as much of the variation in phenotype as possible. The quality of models was assessed using residual plots. Models were fitted to the percentage of the maximum possible AUDPC, but if the residual plots indicated non-normality or heteroscedasticity, AUDPC was transformed by the empirical logit transformation using the smallest possible AUDPC value as a coefficient to avoid logarithms of zero55. Generally, damage data were analysed on the percentage scale, whereas pycnidial coverage usually required logit transformation. Mean pycnidia and damage scores for each genotype were estimated through the R package emmeans56. Statistical analysis used R57 v.4.2.2.
GWAS from the Watkins collection
The markers used for GWAS of the Watkins collection were ~10 Mb core SNPs generated from whole-genome shotgun sequencing of accessions and alignment to Chinese Spring. Extreme outlier values of phenotypic data were removed. We calculated the kinship matrix as the covariate using GEMMA-kin. On this basis, we performed GWAS using GEMMA (v.0.98.1), a computationally efficient method of conducting GWAS on large datasets58, with the command "gemma -miss 0.9 -gk -lm -k kinship.txt -maf [num]" (num: value of minor allele frequency threshold). In-house R scripts were used to visualize associations between SNPs and log pAUDPC as −log10 transformed P values from Wald statistics, which follow a χ2 distribution under the null hypothesis of no association. Multiple test corrections were not applied because the high covariance between closely linked SNPs makes such tests excessively conservative. Stb6 and Stb15 were identified as peaks of −log10(P) in the regions where they are known to be located3,17,19, and STBWat1 was identified as a prominent new peak.
Estimation of haplotypes/alleles of candidate genes
A Python script was written to identify haplotypes of the six candidate Stb genes in the 6AS locus (https://github.com/ambernhafeez/VCFparse_distance.git). The script parsed variant call format files (VCFs) from the alignment of Watkins and wheat lines to Chinese Spring (see above). This produced a matrix of SNP distance between all accessions, which was used to determine haplotype groups. The R package pheatmap59 (v1.0.12) was used to generate heat maps arranged in dendrograms from distance matrices, including associated phenotype data. Iterations of the VCF parsing script were run and plotted to identify the most useful variation for haplotype calling. Ultimately, the whole gene sequence was analysed (rather than, for example, exons alone). The dendrogram produced was manually analysed to estimate the number of haplotype groups present, defined by having a very similar number of SNPs different from Chinese Spring (SNP distance). Clusters were estimated using the cutree function in pheatmap; several iterations were performed to determine the number of clusters/haplotypes that were most informative, particularly for explaining phenotypes. This method was used to determine which Watkins landraces carry the functional ArinaLrFor allele of the Stb15 candidate as well as the functional Chinese Spring allele of Stb6: haplotype groups that contained the resistant accession and were associated with resistance were most likely to represent the functional allele.
Generation of figures presenting Stb gene alleles in the Watkins collection
Figures wherein Stb gene alleles were plotted (Figs. 1a and 3a,b) were generated in R using ggplot2 (ref. 60, v.3.5.1) and cowplot61 (v.0.7.0). For Fig. 3a, the R package ggmap62 (v.3.0.0) was implemented for generating the map and plotting coordinates.
A principal component analysis was conducted in R v.4.2.2 using the package vcfR63 (v.1.14.0) to process the WatSeq VCF data for Stb15, the base R prcomp function to compute the principal component analysis and the vegan64 package (v.2.5-7) for further analysis. Composite main figures and illustrations were generated in Adobe Illustrator 2023.
Identification of candidate Stb genes by bioinformatics
Candidate STB resistance genes were identified by selecting the most likely candidate from the genes in the LD block most highly associated with STB response. Several factors were considered, such as the SNP P value (for association with STB response), gene class, the presence of differential SNPs between susceptible and resistant wheat varieties, and the strength of correlation of predicted resistant haplotypes with STB responses (described above).
To further confirm the Stb15 candidate, a second iteration of the GWAS was run with lines carrying predicted functional alleles of the candidate gene removed. This resulted in the loss of the association of the 6AS locus with resistance, implying that the lines removed did contain the 6AS resistance.
Generation of an Arina EMS population
The generation of the Arina EMS population is described in Kolodziej et al.48. EMS mutagenesis of cv. Arina was performed with concentrations of 0.6% or 0.45% EMS (Sigma Aldrich). Seeds were incubated for 16 h in water at 4 °C, dried for 8 h on filter paper and incubated for 16 h with shaking at room temperature in EMS solution. After being washed three times for 30, 45 and 60 min and for another 30 min under running tap water, the seeds were pre-germinated on humid filter paper. Three thousand seeds of BC2F5-85 were mutagenized, and the pre-germinated seeds were propagated in the field. Single spikes of M0 plants were harvested, and M1 plants were grown and harvested in the field.
Validation of the Stb15 candidate through screening an Arina EMS population
When available, 12 seeds per M2 family were sown in a mixture of 1/2 blond and 1/2 brown peat mosses (Humustar soil, NPK 14-16-18, SARL Activert) and kept at 6 °C for four days. Subsequently, the plants were grown in a CER with sodium lamps (HQI-TS 250 W/D UVS FC2 FLH1; intensity, 300 µmol m−2 s−1) with 16 h, 21 °C day / 8 h, 18 °C night conditions and a relative humidity of 85%. Fourteen days after sowing, the plants were inoculated with a hand sprayer (Elyte 2, Berthoud) with Z. tritici isolate IPO88004. The plants were covered with plastic bags for three days before being returned to normal conditions. Visual evaluations were conducted at 21 and 28 days post-inoculation. All M2 plants carrying pycnidia were self-pollinated. M3 plants were evaluated for resistance to isolate IPO88004, following the procedure described above, except that inoculations were performed on six-centimetre sections in the middle of the second leaf using a paintbrush. Three plants per M3 family were inoculated in two independent experiments. The Z. tritici inoculum was prepared using YG and YPD media following the procedure described in Battache et al.28. Inoculation with concentrations of 1 × 106 spores per ml and 1 × 107 spores per ml, supplemented with 0.05% (v/v) Tween-20, were used for inoculating M2 and M3 plants, respectively. The Stb15 candidate gene was sequenced from each susceptible M3 plant using Sanger sequencing following PCR amplification using the primer pairs Stb15F1/Stb15R1 and Stb15F3/Stb15R3 and the Phusion High-Fidelity Master Mix.
The primers were as follows:
Stb15F1: TCCTACTACTAGCCAAGCATGTC
Stb15R1: GCCATTGCCGTTAGAAACAG
Stb15F3: CTGTTCGAGGGAGGTTCCTA
Stb15R3: GTGCAAAGACCGCAGTATGT
Design of the Stb15 binary vector construct
A wheat transformation vector was assembled using standard Golden Gate MoClo assembly65 and traditional digestion and ligation cloning. The level 1 plasmids pL1P1R PvUbiP::hpt–int::35sT selection cassette, pICH47742 L1P2 multiple cloning site and LacZ (Addgene no. 48001), and pL1P3ZmUbiP::GRF–GIF::NosT (Addgene no. 198047) were assembled into the level 2 acceptor pGoldenGreenGate-M (pGGG-M) (Addgene no. 165422) binary vector66 along with end linker pELE-3 (Addgene no. 48018). The resulting plasmid was named pGGG L2 PvUH GGLacZ GRF–GIF (available as Addgene no. 226630). The Stb15 gene sequence was analysed using the software Geneious Prime v.2020.2.4 (Biomatters), and two restriction enzymes (SbfI and SacI) were chosen for digestion/ligation cloning. The sequence containing the Stb15 gene (6,077 bp), consisting of a 1,917-bp promoter, a 136-bp 5′ untranslated region, a 2,290-bp coding sequence, a 306-bp 3′ untranslated region and a 1,404-bp terminator, was synthesized (Invitrogen, Thermo Fisher Scientific) with restriction enzyme recognition sites SbfI and SacI added to the 5′ and 3′ ends, respectively. The Stb15 gene synthon was cloned into pGGG L2 PvUH GGLacZ GRF–GIF within the multiple cloning site using SbfI and SacI digestion/ligation. The resulting plasmid was named pGGG L2 TaStb15 and was electroporated into the hypervirulent Agrobacterium strain AGL1 (refs. 67,68).
Agrobacterium transformation of T. aestivum cv. Fielder
The wheat transformation method described here was based on an existing method69 with slight modifications. The construct incorporated GRF4–GIF1 technology70. Briefly, wheat cv. Fielder was grown in a CER under a long-day photoperiod (16 h at 600 μmol m−2 s−1 light, at 20 °C day and 16 °C night). Wheat spikes were collected ~14 days post anthesis (early milk stage GS73) when the immature embryos were 1–1.5 mm in diameter. Under aseptic conditions, immature embryos were isolated from surface-sterilized grain.
The isolated immature embryos were pretreated by centrifugation in liquid medium prior to Agrobacterium inoculation. The embryos were transferred to co-cultivation medium, scutellum side up, and incubated at 24 °C in the dark for three days of co-cultivation. The embryogenic axes were excised and discarded; then, the embryos were transferred to wheat callus induction (WCI) medium without selection for five days at 24 °C in the dark. After five days, the embryos were transferred to WCI containing 15 mg l–1 hygromycin and incubated at 24 °C in the dark. Subculturing onto fresh WCI with hygromycin selection at 15 mg l–1 occurred every two weeks over a five-week period. For the final, fifth week on WCI, the cultures were maintained in low-light conditions at 24 °C and then transferred onto wheat regeneration medium supplemented with 0.5 mg l–1 zeatin and 15 mg l–1 hygromycin in deep Petri dishes (90 mm diameter × 20 mm) and cultured under fluorescent light (100 μM m−2 s−1) with a 16 h photoperiod. Regenerated plantlets were transferred to De Wit culture tubes (Duchefa-Biochemie, W1607) containing rooting medium supplemented with 20 mg l−1 hygromycin. After approximately ten days, rooted plants were transferred to soil (John Innes cereal mix in 24-cell trays) and acclimatized68. The transgenic plants were maintained under the same growing conditions as donor material with a long-day photoperiod (16 h at 600 μmol m−2 s−1 light, at 20 °C day and 16 °C night). Transgenesis was confirmed and transgene copy number analysis performed on 58 independent lines using a Taqman relative quantification assay. The relative values of the hygromycin resistance gene (hpt) and CO2 (Constans-like, AF490469) were compared in a multiplexed reaction using specific probes and primers68. Relative quantification values (ΔΔCt) were calculated to determine transgene copy number71.
Experimental validation of the Stb15 transgenics
The experiment to test the effect of the Stb15 candidate on STB symptoms caused by the Stb15-avirulent isolate IPO88004 and the virulent control isolate IPO92006 was sown in 40-well seed trays with two experimental replicates per tray in a randomized design (ten replicates per line). This assay was then repeated with a new randomized design in a separate trial, giving up to 20 replicates per line for IPO88004 and 10 replicates per line for IPO92006. The conditions and infection protocol were as described above under ‘Design and infection protocol for pathology assays at the JIC’, except the inoculum concentration of IPO88004, which was adjusted to 107 spores per ml because of the inherent partial resistance of cv. Fielder. The percentage leaf area covered by lesions containing pycnidia was analysed by linear mixed modelling as described in the section ‘Statistical analysis of STB phenotypes’ above. The random-effect model for pycnidia data was Replicate within Tray within Test, and Tray within Test and the fixed-effects model was Test × Isolate + Isolate + Isolate × Variety. For damage, the same fixed-effects model was used, but the random effect of Test within Variety was included due to its significant explanation of residual variance as determined by nested deviance tests. The R package emmeans was used to perform Tukey-honest-significant-difference-adjusted pairwise comparisons between all lines.
Identification of intron/exon structure of Stb15
Prepublication access to an updated genome annotation of ArinaLrFor based on RNAseq data was provided by A. Hall and M. Spannagl72. Additionally, alignments of the RNAseq data were manually checked in IGV (v.2.14.0)73 to confirm the intronless gene structure of Stb15.
Protein structure prediction of Stb15
The protein structure encoded by the Stb15 candidate gene in Arina was predicted using AlphaFold (v.2.3.2)74. The quality of the highest-confidence prediction was assessed using AlphaFold Analyser (v.2.0.0). Each protein sequence was also annotated using InterProScan75 (v.5.61-93.0). These annotations were visualized on the high-confidence protein structure using PyMol76 (v.2.5.2), and domain boundaries were manually expanded upon to include unannotated amino acids.
KASP genotyping of European cultivars
KASP genotyping was carried out as described in Saintenac et al.4 on 278 European wheat cultivars. The marker sequences were as follows:
F = GAAGGTGACCAAGTTCATGCTGGTTTCAACTTGCAATATGATC
V = GAAGGTCGGAGTCAACGGATTGGTTTCAACTTGCCATATGATT
C = AGTGAACCAGGTGCCAAAAC
Analyses of sequence evolution
Identification of potential Stb15 homologues in plants
High-quality reference genome protein annotations of plant species were downloaded (Supplementary Table 16). Local BLASTp databases were generated using command-line BLAST77 v.2.13.0. The amino acid sequence of the functional Stb15 allele from ArinaLrFor was used as a query sequence for BLAST searches against protein databases of each species. The top 30 hits were recovered to ensure that no potential orthologues or paralogues were missed.
Protein alignment
A protein alignment was generated using MUSCLE78 v.3.8.31 with the default settings. Sequences were removed if they contained large (>350 amino acids) and divergent insertions that disrupted the alignment or if they contained less than two of the domains present in the ArinaLrFor Stb15 sequence. Multiple splice variants were included if their predicted amino acid sequence varied.
Phylogenetic tree construction and analysis of the Stb15 clade
ModelFinder79 was used to predict the best evolutionary model for the alignment (JTT + R10) implemented via IQ-TREE80 v.1.6.10. Branch supports were obtained with ultra-fast bootstrap (UFBoot2 (ref. 81)), and tree reconstruction was performed using IQ-TREE. The least-repetitive clade containing Stb15 was extracted, and sequence conservation was analysed in Geneious v.2022.2.2. Genome annotations (GFFs) for each orthologue were used to draw gene structures in R v.4.2.2. For Fig. 3c and Supplementary Fig. 5, all exon annotations for each gene are presented within a single leaf, and splice variants were pruned from the tree. The tree image was generated using iTOL82 v.6.
Alignment for the consensus sequence of the inner Stb15 clade
A small alignment of the inner Stb15 clade was generated by MUSCLE within Geneious v.2022.2.2. It was noticed that the Zea mays homologue Zea_mays_Zm00001eb119590_P002 was in fact a tandem duplication containing two identical sequences of a protein encoding a partial BTL and full SLG, PAN and S/TPK domains. To reduce disruption of the alignment, one half of this protein sequence was retained. A screenshot of the consensus chart from the Geneious alignment was used in Fig. 3d.
Microsynteny analysis of Stb15 orthologues in T. aestivum, Aegilops tauschii, T. durum and Hordeum vulgare
Genome annotation files (GFFs) were used to extract the names and coordinates of ten genes on each side of the Stb15 homologues identified above, from the same genome assemblies (see ‘Analyses of sequence evolution’). Protein sequences were extracted from peptide files associated with the genome assemblies used or exported from Ensembl Plants, and the NCBI BLAST server was used to conduct a BLASTp search. Other proteins with high similarity to the query and the Conserved Domains results were used to identify the protein family. If informative results were not gained in this way, InterProScan was also used. If protein sequences were truncated or uninformative, a tBLASTx search of the nucleotide sequence was conducted against T. aestivum, Brachypodium distachyon and Oryza sativa. In some cases, such as Triticum urartu and Aegilops searsii, synteny with ArinaLrFor could not be found for the investigated genes. This analysis was not exhaustive as it focused on a small number of genes within a small genomic region. It was noticed that a truncated protein was predicted for the ArinaLrFor 6B orthologue of Stb15 despite high similarity to the nucleotide sequence of Stb15. This is why this gene appears in the microsynteny analysis but not the phylogenetic trees of proteins with similarity to Stb15 (Fig. 3c and Supplementary Fig. 8). The figure depicting these results (Supplementary Fig. 9) was generated with Inkscape v.1.3.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The SNP data5 are available via GrassRoots at https://opendata.earlham.ac.uk/wheat/under_license/toronto/WatSeq_2023-09-15_landrace_modern_Variation_Data/WatSeq_VCF_ChineseSpringRefSeqv1.0/. The pathology data are available via Zenodo at https://doi.org/10.5281/zenodo.14515753 (ref. 83). The Stb15 sequence has been reported with an annotated transcript on Ensembl Plants (TraesARI6A03G03215890.1).
Code availability
Customs scripts for analysis of STB pathology data, plot generation, haplotype analysis and principal coordinates analysis are available via GitHub at https://github.com/ambernhafeez/Stb15_Project_Analyses. AlphaFold Analyser is available via GitHub at https://github.com/Orpowell/alphafold-analyser. Software for SNP mapping and GWAS-QTL mapping5 and plotting is available via GitHub at https://github.com/ShifengCHENG-Laboratory/WWWG2B and https://github.com/JIC-CSB/WatSeqAnalysis/.
References
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
O’Driscoll, A., Kildea, S., Doohan, F., Spink, J. & Mullins, E. The wheat–Septoria conflict: a new front opening up? Trends Plant Sci. 19, 602–610 (2014).
Saintenac, C. et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50, 368–374 (2018).
Saintenac, C. et al. A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against Septoria tritici blotch. Nat. Commun. 12, 433 (2021).
Cheng, S. et al. Harnessing landrace diversity empowers wheat breeding. Nature 632, 823–831 (2024).
Stukenbrock, E. H., Banke, S., Javan-Nikkhah, M. & McDonald, B. A. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 24, 398–411 (2007).
Hafeez, A. N. et al. Creation and judicious application of a wheat resistance gene atlas. Mol. Plant 14, 1053–1070 (2021).
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Stotz, H. U., Mitrousia, G. K., de Wit, P. J. G. M. & Fitt, B. D. L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500 (2014).
Cook, D. E., Mesarich, C. H. & Thomma, B. P. H. J. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563 (2015).
Kanyuka, K. & Rudd, J. J. Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8 (2019).
Kema, G. H. J., Yu, D., Rijkenberg, F. H. J., Shaw, M. W. & Baayen, R. P. Histology of pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology 7, 777–786 (1996).
Shaw, M. W. Assessment of upward movement of rain splash using a fluorescent tracer method and its application to the epidemiology of cereal pathogens. Plant Pathol. 36, 201–213 (1987).
Brown, J. K. M., Chartrain, L., Lasserre-Zuber, P. & Saintenac, C. Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 79, 33–41 (2015).
Yang, N., Mcdonald, M. C., Solomon, P. S. & Milgate, A. W. Genetic mapping of Stb19, a new resistance gene to Zymoseptoria tritici in wheat. Theor. Appl. Genet. 131, 2765–2773 (2018).
Langlands-Perry, C. et al. Resistance of the wheat cultivar ‘Renan’ to Septoria leaf blotch explained by a combination of strain specific and strain non-specific QTL mapped on an ultra-dense genetic map. Genes 13, 100 (2022).
Brading, P. A., Verstappen, E. C. P., Kema, G. H. J. & Brown, J. K. M. A gene-for-gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology 92, 439–445 (2002).
Ghaffary, S. M. T. et al. New broad-spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 124, 125–142 (2012).
Arraiano, L. S. et al. A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola. Plant Pathol. 56, 73–78 (2007).
Arraiano, L. S. & Brown, J. K. M. Identification of isolate-specific and partial resistance to septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 55, 726–738 (2006).
Wingen, L. U. et al. Establishing the A. E. Watkins landrace cultivar collection as a resource for systematic gene discovery in bread wheat. Theor. Appl. Genet. 127, 1831–1842 (2014).
Winfield, M. O. et al. High-density genotyping of the A.E. Watkins Collection of hexaploid landraces identifies a large molecular diversity compared to elite bread wheat. Plant Biotechnol. J. 16, 165–175 (2018).
Ajaz, S., Benbow, H. R., Christodoulou, T., Uauy, C. & Doohan, F. M. Evaluation of the susceptibility of modern, wild, ancestral, and mutational wheat lines to Septoria tritici blotch disease. Plant Pathol. 70, 1123–1137 (2021).
Arora, S. et al. A wheat kinase and immune receptor form host-specificity barriers against the blast fungus. Nat. Plants 9, 385–392 (2023).
Kema, G. H. J. et al. Variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem I. Interactions between pathogen isolates and host cultivars. Phytopathology 86, 213–220 (1996).
Chartrain, L., Sourdille, P., Bernard, M. & Brown, J. K. M. Identification and location of Stb9, a gene for resistance to septoria tritici blotch in wheat cultivars Courtot and Tonic. Plant Pathol. 58, 547–555 (2009).
Amezrou, R. et al. A secreted protease-like protein in Zymoseptoria tritici is responsible for avirulence on Stb9 resistance gene in wheat. PLoS Pathog. 19, e1011376 (2023).
Battache, M. et al. Blocked at the stomatal gate, a key step of wheat Stb16q-mediated resistance to Zymoseptoria tritici. Front. Plant Sci. 13, 921074 (2022).
Sun, Y., Qiao, Z., Muchero, W. & Chen, J. G. Lectin receptor-like kinases: the sensor and mediator at the plant cell surface. Front. Plant Sci. 11, 1989 (2020).
Avni, R. et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 110, 179–192 (2022).
Bartoli, C. & Roux, F. Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front. Plant Sci. 8, 763 (2017).
Chartrain, L., Brading, P. A. & Brown, J. K. M. Presence of the Stb6 gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in cultivars used in wheat-breeding programmes worldwide. Plant Pathol. 54, 134–143 (2005).
Steuernagel, B. et al. The NLR-Annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. https://doi.org/10.1104/pp.19.01273 (2020).
Wulff, B. B. H., Chakrabarti, A. & Jones, D. A. Recognitional specificity and evolution in the tomato–Cladosporium fulvum pathosystem. Mol. Plant Microbe Interact. 22, 1191–1202 (2009).
Thomas, C. M., Dixon, M. S., Parniske, M., Golstein, C. & Jones, J. D. G. Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Phil. Trans. R. Soc. B 353, 1413–1424 (1998).
Larkan, N. J. et al. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 104, 892–900 (2020).
Larkan, N. J. et al. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605 (2013).
Miyakawa, T. et al. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 166, 766–778 (2014).
Burton, R. A. & Fincher, G. B. Evolution and development of cell walls in cereal grains. Front. Plant Sci. 5, 456 (2014).
Singh, P. & Zimmerli, L. Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 4, 124 (2013).
Wang, Y. et al. Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5, 1129–1135 (2019).
van der Hoorn, R. A. L. & Kamoun, S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017 (2008).
Ma, L. S. et al. Maize antifungal protein AFP1 elevates fungal chitin levels by targeting chitin deacetylases and other glycoproteins. mBio 14, e00093-23 (2023).
Bouwmeester, K. et al. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 7, e1001327 (2011).
Amezrou, R. et al. Quantitative pathogenicity and host adaptation in a fungal plant pathogen revealed by whole-genome sequencing. Nat. Commun. 15, 1933 (2024).
Zhong, Z. et al. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 214, 619–631 (2017).
Kema, G. H. J. et al. Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat. Genet. 50, 375–380 (2018).
Kolodziej, M. C. et al. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nat. Commun. 12, 956 (2021).
Kema, G. H. J. & Van Silfhout, C. H. Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem III. Comparative seedling and adult plant experiments. Phytopathology 87, 266–272 (1997).
Kema, G. H. J. et al. Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem I. Interactions between pathogen isolates and host cultivars. Phytopathology 86, 200–212 (1996).
Kema, G. H., Sayoud, R., Annone, J. G. & Van Silfhout, C. H. Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem II. Analysis of interactions between pathogen isolates and host cultivars. Phytopathology 86, 213–220 (1996).
Patterson, H. D. & Williams, E. R. A new class of resolvable incomplete block designs. Biometrika 63, 83–92 (1976).
Arraiano, L. S., Brading, P. A. & Brown, J. K. M. A detached seedling leaf technique to study resistance to Mycosphaerella graminicola (anamorph Septoria tritici) in wheat. Plant Pathol. 50, 339–346 (2001).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. https://doi.org/10.18637/jss.v082.i13 (2017).
McGrann, G. R. D. et al. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. J. Exp. Bot. 65, 1025–1037 (2014).
Lenth, R. V. et al. emmeans: Estimated marginal means, aka least-squares means. R package version 1.10 https://doi.org/10.32614/CRAN.package.emmeans (2023).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012).
Kolde, R. pheatmap: Pretty heatmaps. R package version 1.0.12 (2012).
Wickham, H. Package ‘ggplot2’: Elegant Graphics for Data Analysis (Springer, 2016).
Wilke, C. O. cowplot: Streamlined plot theme and plot annotations for ‘ggplot2’. R package version 0.7.0 https://CRAN.R-project.org/package=cowplot (2016).
Kahle, D. & Wickham, H. ggmap: spatial visualization with ggplot2. R J. 5, 144–161 (2013).
Knaus, B. J. & Grünwald, N. J. vcfr: a package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53 (2017).
Oksanen, J. et al. vegan: Community ecology package. R package version 2.5-7 (2020).
Werner, S., Engler, C., Weber, E., Gruetzner, R. & Marillonnet, S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3, 38–43 (2012).
Smedley, M. A., Hayta, S., Clarke, M. & Harwood, W. A. CRISPR–Cas9 based genome editing in wheat. Curr. Protoc. 1, e65 (2021).
Lazo, G. R., Stein, P. A. & Ludwig, R. A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N. Y.) 9, 963–967 (1991).
Hayta, S. et al. An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods 15, 152 (2019).
Hayta, S., Smedley, M. A., Clarke, M., Forner, M. & Harwood, W. A. An efficient Agrobacterium-mediated transformation protocol for hexaploid and tetraploid wheat. Curr. Protoc. 1, e58 (2021).
Debernardi, J. M. et al. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279 (2020).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
White, B. et al. De novo annotation of the wheat pan-genome reveals complexity and diversity within the hexaploid wheat pan-transcriptome. Preprint at bioRxiv https://doi.org/10.1101/2024.01.09.574802 (2024).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res. 51, D418–D427 (2023).
The PyMol Molecular Graphics System v. 2.5. (Schrodinger LLC, 2021).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Hafeez, A. N. et al. Stb15 pathology data, Zenodo, https://zenodo.org/records/14515753 (2025).
International Wheat Genome Sequencing Consortium Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Acknowledgements
We thank C. Uauy, A. Hall and M. Spannagl for prepublication access to ArinaLrFor RNAseq data and associated genome annotations; T. O’Hara for sharing resources; T. Olsson for Python training and consultation; S. Ghosh and G. Radhakrishnan for advice on phylogenetics; and the JIC Germplasm Resources Unit, a National Bioscience Research Infrastructure supported by BBSRC (grant no. BBS/E/JI/23NB0001), for conserving and supplying germplasm through www.seedstor.ac.uk. This research was supported by the NBI Research Computing group, the Informatics Platform and Horticultural Services at the JIC, UK, and the experimental infrastructure VégéPôle, INRAE, France. Funding was provided by the Biotechnology and Biological Sciences Research Council through a Norwich Research Park Doctoral Training Programme studentship (A.N.H.) and Institute Strategic Programmes on Plant Health (BBS/E/J/000PR9798; J.K.M.B.), Advancing Plant Health (BB/X010996/1; J.K.M.B.), Designing Future Wheat (BBS/E/J/000PR9780; S.G., L.U.W. and B.B.H.W.) and Delivering Sustainable Wheat (BB/X011003/1; S.G.); King Abdullah University of Science and Technology (B.B.H.W.); the National Key Research and Development Program of China (2023YFF1000100 and 2023YFA0914600; S.C.); the EU Horizon 2020 Marie Skłodowska-Curie programme (674964; B.K.); and the Swiss National Science Foundation (310030_204165; B.K.).
Author information
Authors and Affiliations
Contributions
The authors are listed in alphabetical order by last name apart from the first three and last four authors. The Z. tritici pathology work on Watkins landraces was planned and implemented by A.N.H., L.C. and R.K. with supervision from J.K.M.B. Statistical analysis of the pathology data was carried out by A.N.H. and J.K.M.B. Prepublication collaboration (within WatSeq), access to genotyping data and the application of GWAS to the project were facilitated primarily by S.C. alongside S.G. and implemented by L.U.W., C.F., W.X. and M.J., as were the generation of the associated figures and the suggestion of candidate genes. A.N.H. and B.S. analysed the WatSeq genotypes for allele characterization and the identification of candidate genes. The cloning strategy was conceived and planned by B.B.H.W. An Arina EMS mutant population was generated by M.C.K. in the labs of B.K. Large-scale screens were planned and implemented by C.S. and F.C., along with the identification and resequencing of induced susceptible mutants and the provision of phenotype images. O.R.P. generated and annotated the AlphaFold model of Stb15. M.A.S. designed a vector carrying Stb15, which was transformed into wheat by S.H. and M.C., followed by single-seed descent to T2 and screening with Z. tritici by L.C. and A.N.H. and statistical analysis by J.K.M.B. and A.N.H. The study of Stb15 homologues and gene structural variation was conducted by A.N.H. The composite figures were designed by A.N.H., B.B.H.W. and J.K.M.B. and generated by A.N.H. A.N.H. drafted the manuscript with extensive input and revisions from B.B.H.W., J.K.M.B. and C.S. and contributions from B.K., M.C.K., L.C., S.H., M.A.S., C.F. and M.J.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Tzion Fahima, Yinghui Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Tables 1–17 and Text 1.
Supplementary Tables
Spreadsheets with data for Supplementary Tables 4, 11, 12 and 15–17.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hafeez, A.N., Chartrain, L., Feng, C. et al. Septoria tritici blotch resistance gene Stb15 encodes a lectin receptor-like kinase. Nat. Plants 11, 410–420 (2025). https://doi.org/10.1038/s41477-025-01920-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-01920-2