Abstract
The waxy cuticle layer is crucial for plant defence, growth and survival, and is produced by epidermal cells, which were thought to be specified only during embryogenesis. New surface cells are exposed during abscission, by which leaves, fruits, flowers and seeds are shed. Recent work has shown that nonepidermal residuum cells (RECs) can accumulate a protective cuticle layer after abscission, implying the potential de novo specification of epidermal cells by transdifferentiation. However, it remains unknown how this process occurs and what advantage this mechanism may offer over the other surface protection alternative, the wound healing pathways. Here we followed this transdifferentiation process with single-cell RNA sequencing analysis of RECs, showing that nonepidermal RECs transdifferentiate into epidermal cells through three distinct stages. During this vulnerable process, which involves a transient period when the protective layer is not yet formed, stress genes that protect the plant from environmental exposure are expressed before epidermis formation, ultimately facilitating cuticle development. We identify a central role for the transcription factor MYB74 in directing the transdifferentiation. In contrast to alternative protective mechanisms, our results suggest that de novo epidermal specification supports the subsequent growth of fruit at the abscission site. Altogether, we reveal a developmental programme by which plants use a transdifferentiation pathway to protect the plant while promoting growth.
Similar content being viewed by others
Main
The epidermis is a plant’s first line of defence to safeguard internal tissues1,2,3. In terrestrial plants, the epidermis has evolved to meet the demands of life on dry land by forming a waxy cuticle on its surface, thus protecting the plants from dehydration, pathogens and toxic molecules3. The epidermis also functions as a conduit for conveying environmental cues to the interior of the plant and assists in the differentiation of internal tissues and plant growth4,5,6,7,8. For example, epidermal cells of the shoot apical meristem can modulate their division patterns in response to mechanical stimuli9. Disruptions in epidermal differentiation or cuticle development result in vulnerability to external harm as well as deformed organs and altered plant growth1,3,10,11. When epidermal cells are damaged by abiotic or biotic injuries, plants can use wound-induced suberization as an alternative protection mechanism on damaged, bare surfaces to rapidly prevent pathogen invasion and tissue dehydration12,13,14,15. As the scars are healed, these areas experience lignification of cell walls for further surface protection, but this process impairs future organ expansion and growth12,13.
Organ abscission is a pivotal process by which leaves, fruits, flowers and seeds are shed in response to specific developmental programmes or environmental cues16. Abscission results in the exposure of nonepidermal cells at the surface where the organ was attached, known as the receptacle. Establishing a protective layer at the newly formed surface was originally thought to involve suberin and lignin, as in the case of wound healing17,18,19. However, non-epidermal residuum cells (RECs), located deep in the abscission zone (AZ) of flower receptacles, were observed to accumulate a protective cuticle layer after the abscission of floral organs in Arabidopsis20. Given that cuticle formation is a defining feature of epidermal cells21, this observation suggests that epidermal cells are specified de novo from RECs and direct cuticle biosynthesis during abscission and/or post-abscission. However, it remains unclear how this process occurs and what advantage this mechanism may offer over the wound healing pathways that assemble suberin and lignin to protect the newly exposed surface22,23. In this study, we directly follow the de novo specification of epidermal cells in the AZ and characterize transcriptomic changes that occur throughout this developmental programme at single-cell resolution. We identify MYB74 as a transcription factor that facilitates the process of de novo specification of epidermal cells. Importantly, our results suggest that, contrary to wound healing mechanisms that may restrict future growth at the site, the epidermal transdifferentiation programme contributes to fruit growth.
Results
Three distinct states during REC transdifferentiation
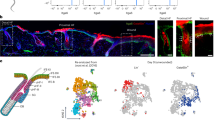
To examine cellular and molecular changes in RECs undergoing transdifferentiation, we conducted single-cell RNA sequencing (scRNA-seq) analysis using REC protoplasts obtained from the AZ of transgenic plants harbouring prEXL1:nls-GFP. EXORDIUM-LIKE 1 (EXL1) is expressed in AZ cells from flower development stage 15 to 17 AZ cells (Fig. 1a). Thus, the transgenic plants expressing prEXL1:nls-GFP can provide information on cellular processes before and after abscission24,25. Protoplasts were, therefore, prepared from these AZ cells and subjected to fluorescence-activated cell sorting as previously described26. Approximately 100,000 GFP-positive protoplasts were immediately subjected to scRNA-seq analysis (Extended Data Fig. 1a). We profiled a total of 2,823 RECs that passed our quality control criteria, excluding secession cells, guard cells and vasculature cells (Extended Data Figs. 1b,c and 2a–d). Unsupervised clustering analysis classified 2,823 cells into five clusters (Extended Data Fig. 2a), which were assigned to three developmental states based on inter-cluster similarity (Fig. 1b and Extended Data Fig. 2e–h). Both pseudotime and RNA velocity analysis consistently predicted a linear differentiation trajectory from states 1 to 3 (Fig. 1b). To gain insights into changing cellular attributes across the three cell states, we performed a label transfer analysis between our scRNA-seq data and publicly available single-cell data from Arabidopsis leaves27 (Fig. 1c and Extended Data Fig. 2i). Our analysis revealed that state 1 RECs primarily have mesophyll cell characteristics. As the cells progress to the transition state, the RECs in state 2 show a diverse range of cellular characteristics found in four distinct cell types: guard cells, mesophyll cells, vascular cells and epidermal cells. The mixed cellular traits observed in cell state 2 suggest that these cells are undergoing a process of developmentally programmed differentiation. In state 3, the majority of cells possess epidermal cell traits.
a, Expression of prEXL1:nls-GFP in the AZ of flowers at stage 15 (S15) to 17 (S17). Floral stages are defined according to previous studies24,25, and S17 indicates stages beyond S16. S15e and S15l indicate early and late S15, respectively. S17e, S17m and S17l indicate early, middle and late S17, respectively (left). Floral organ separation is completed after S15l (marked by an arrow). The positions of stamen (st), petal (p) and sepal (se) AZs are indicated in the bottom rightmost receptacle image. Scale bars, 50 μm. Confocal laser scanning microscope images show the expression of prEXL1:nls-GFP in RECs from longitudinally or transversely sectioned receptacles at S17 (right). Scale bars, 50 μm. b, Developmental progression of de novo specification of epidermal cells analysed by scRNA-seq of RECs shown in a. FA plots were used to visualize cell–cell relationships through a force-directed graph layout. In this layout, cells with similar transcriptional profiles are positioned closer together. The FA plot shows RECs, coloured according to their three developmental states, with RNA velocity inferred by scVelo (left). The FA plot is coloured based on Palantir pseudotime (right). The arrows indicate cellular trajectories inferred from known gene expression patterns (Extended Data Fig. 2). c, UMAP plot showing the leaf cell atlas from a previous study27, which was used for label transfer analysis (top left). FA plot showing the distribution of inferred REC cell types using label transfer analysis (bottom left). Relative abundance of predicted cell types via label transfer in different REC developmental states, revealing that the inferred non-epidermal RECs progress to an intermediate state and undergo transdifferentiation into epidermal cells (right). The same colour code is used for cell types. d, GO enrichment analysis reveals cellular events during REC transdifferentiation from nonepidermal to epidermal cells. Colours representing cell states are consistent with those in b. P values were determined using one-sided hypergeometric tests followed by multiple-testing correction with the g:SCS method from g:Profiler. e, Dot plot showing the expression of marker genes for the three states of RECs during transdifferentiation.
To gain further insights into the activities of genes being expressed at each state, we performed Gene Ontology (GO) enrichment analysis of the state-specific genes (Fig. 1d and Extended Data Fig. 3a,b). We found that genes associated with translation and photosynthesis were highly expressed in the state 1 RECs and subsequently downregulated in states 2 and 3 (Fig. 1d,e). One feature of epidermal cells, though species dependent, is the lack of photosynthetic chloroplasts28. To validate this finding in vivo, we analysed the expression of three photosynthesis-related genes in transgenic plants. Longitudinal- and transverse-section drawings illustrate the cell types in the AZ (Fig. 2a). Longitudinal sections of the receptacles show that the expression of RUBISCO ACTIVASE (RCA), CHLOROPHYLL A/B BINDING PROTEIN 1 (CAB1) and RUBISCO SMALL SUBUNIT 1A (RBCS1A) was detected in all AZ cells before abscission (flower stage 15) and disappeared in the outermost cells in the AZ after the completion of abscission (Fig. 2b). This result implies that nonepidermal RECs acquire traits associated with epidermal cells.
a, Longitudinal- and transverse-section drawings of the cell types in the AZ. eRECs, RECs with epidermal fate. b, Confocal images of longitudinally sectioned receptacles show the anatomy of the AZs at floral stages 15–17 (top to bottom). Spatiotemporal expression of the photosynthesis-related genes (RCA, CAB1, RBCS1A) and the stress response genes (ABI5, ERF1A) is shown. The red dashed oval indicates the sepal AZ at S15. The cell wall is visualized using calcofluor white staining. Scale bars, 25 µm. c, The progression of cuticle layer formation after abscission is visualized using scanning electron micrographs (top) and the cuticle staining dye auramine O (bottom). The cell wall is visualized by calcofluor white (shown in blue), and the cuticle stained with auramine O is shown in red. The yellow arrowheads indicate the de novo accumulation of cuticles on the newly formed surface, while the blank arrowheads point to the cuticle on the pre-existing epidermal cells. Scale bars, 5 µm (top) and 25 µm (bottom). d, The spatiotemporal expression of epidermal markers ATML1, BDG1 and GPAT4. The Z-projection of surface scans shows the expression of the epidermal marker genes in RECs at S17m. The longitudinal- and transverse-section images show a single layer of de novo specified epidermal cells. Scale bars, 50 µm. e, The expression pattern of MYB74 in multiple cell layers in longitudinally and transversely sectioned AZs of transgenic plants harbouring prMYB74:nls-GFP at floral stages 15–17m. These images are generated by stacking optical sections, regardless of its differential expression level at each stage (the expression dynamics of MYB74 are accurately depicted in Extended Data Fig. 6a,b). Scale bars, 50 µm. f, Co-expression of MYB74 and HAE in multiple cell layers of AZs before and after abscission. Scale bar, 25 µm.
RECs upregulate stress response genes before the vulnerable transition period
After abscission, RECs do not possess a protective layer until a cuticle is formed de novo on their surface20. To visualize the temporal emergence of epidermal establishment in detail, we surveyed the REC surface from stages 16 to 17, during which a cuticle layer accumulates de novo. Scanning electron micrographs revealed unfilled gaps between cells, which were eventually filled with cuticular materials (Fig. 2c). This observation was supported by cuticle deposition, as visualized by auramine O staining29 (Fig. 2c). The temporary absence of the cuticle layer on RECs renders them susceptible to environmental harm, including plant pathogens20. Interestingly, genes associated with biotic and abiotic stress responses were highly enriched and preferentially upregulated in RECs in state 2, indicating that stress response genes are expressed in the RECs ahead of the imminent vulnerable period (Fig. 1d,e).
To validate these dynamic changes in vivo, we examined the expression patterns of the stress response genes30,31, ABA-INSENSITIVE 5 (ABI5) and ETHYLENE RESPONSE FACTOR 1A (ERF1A), in RECs. Transgenic plants harbouring prABI5:nls-GFP or prERF1A:nls-GFP revealed that these two genes are expressed in RECs at stages 15 and 16. Their expression was reduced in the outermost cells by stage 17, when the protective cuticle layer is formed on the surface (Fig. 2b and Extended Data Fig. 4a). As nonepidermal RECs form a new plant surface, they must take on the role of protecting the receptacle and siliques against infection and environmental changes. Given that there is a transient period during which there is no protective cuticle layer on the newly formed epidermal surface20 (Fig. 2c), RECs undergoing transdifferentiation into epidermal cells express genes that ready the plant for potential biotic and abiotic stress before this vulnerable period.
De novo specification of epidermal cells
After the transition state, the RECs progress towards epidermal cells (state 3), and the expression levels of stress response genes decrease. At the same time, we found that the state 3 RECs started to express genes related to cuticle development, fatty acid and wax biosynthesis, and cutin biosynthesis (Fig. 1d,e). The products of these genes are linked to cell surface integrity and protection in accordance with the observed surface (Fig. 2c). We examined the in vivo expression of cuticle development genes using the transgenic plant lines and found that expression of the cutin-biosynthesis genes, BODYGUARD 1 (BDG1) and GLYCEROL-3-PHOSPHATE SN-2-ACYLTRANSFERASE 4 (GPAT4), was observed in outermost RECs (Fig. 2d and Extended Data Fig. 4b,c). In addition to cuticle development-related genes, the epidermal cell marker ATML1 (ref. 10) increased in the outermost cells (Fig. 2d). These data support the hypothesis that nonepidermal RECs undergo transdifferentiation to become epidermal cells. Overall, these results indicate that nonepidermal RECs (state 1) proceed to the transition state (state 2) and gradually acquire the characteristics of epidermal cells (state 3), including the formation of the cuticle layer on the surface.
MYB74, a key factor in REC transdifferentiation
To identify the molecular players that control REC transdifferentiation into epidermal cells, we examined the dynamics of transcription factors (TFs) with cell-state-preferential expression patterns (Extended Data Fig. 5). A schematic drawing in Extended Data Fig. 5a illustrates which stages of fruit development32 correspond to cellular states 1, 2 and 3. We focused on TFs that are preferentially expressed during state 2, which encompasses the transition from the nonepidermal to the epidermal identity. In contrast to the state 3-specific TFs, which are related to epidermal cells and cuticle formation, most of the state 2-enriched TFs are related to stress responses, consistent with the cellular activities in state 2. Nevertheless, MYB7 and MYB74 genes attracted our attention because MYB TFs have previously been implicated in epidermal cuticle formation, trichome development, stomatal patterning and stress responses33,34,35. Of the two MYB genes, MYB74 was of particular interest because the closely related MYB41 is involved in suberin development and cell expansion and acts upstream of several MYBs36,37,38,39,40.
To test the possibility that MYB74 functions in transdifferentiation, we analysed the spatiotemporal expression of MYB74 in RECs from plants carrying prMYB74:nls-GFP-GUS. We discovered that MYB74 starts to appear before abscission completes, reaches its highest expression level during the transition state and slowly vanishes in the AZ cells (Extended Data Fig. 6a,b). Longitudinal and transverse sections of transgenic receptacles show that MYB74 is detected in at least three cell layers in the AZ from stage 15 to mid-17 (Fig. 2e), but is hardly detected in cells from late stage 17 (Extended Data Fig. 6a,b), consistent with transdifferentiation as inferred in the pseudotime analysis. Note that the expression dynamics of MYB74 are accurately depicted in Extended Data Fig. 6a,b. The images in Fig. 2e are generated to show that MYB74 is expressed in multiple layers of cells by stacking optical sections, regardless of its differential expression level at each stage. The receptacle sections also showed that RECs may consist of multiple cell layers. Longitudinal sections of the transgenic plants revealed that BLADE ON PETIOLE2 (BOP2), HAESA (HAE), QUARTET2 (QRT2) and HAESA-LIKE2 (HSL2), genes that are involved in abscission19,41,42, are also expressed in these multiple cell layers (Fig. 2f and Extended Data Fig. 6c). Organ abscission is initiated by the binding of the peptide ligand INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) to the leucine-rich repeat receptor-like kinases HAE and HSL242,43,44. The spatiotemporal expression of MYB74 is not altered in the abscission signalling mutants hae hsl2 and ida (Extended Data Fig. 6d). This result suggests that MYB74 is expressed independently of the IDA-HAE HSL2-mediated abscission signalling and that MYB74 acts in post-abscission events, although we cannot completely exclude the possibility that IDA and/or HAE HSL2 affect MYB74.
MYB74 has been implicated in salt and osmotic stress responses45,46; however, its role in development remains largely unknown. To test whether MYB74 is linked to the transdifferentiation of RECs into epidermal cells, we generated transgenic plants harbouring prUBQ10:XVE>>MYB74 (iMYB74) to ectopically express MYB74 using the β-oestradiol-inducible system47. RNA-seq analysis of the β-oestradiol-treated receptacles at stage 16 to early 17 showed that the induction of MYB74 expression led to a drastic change in the transcriptome, resulting in the identification of 1,763 genes upregulated and 3,175 genes downregulated in RECs (Extended Data Fig. 7a,b and Supplementary Table 1). Similar to the scRNA-seq results, 58 photosynthesis-related genes are substantially downregulated, whereas 95 cuticle-development- and lipid-biosynthesis-related genes are highly upregulated in the plants (Fig. 3a and Extended Data Fig. 7a,c). These results point to a role for MYB74 in the transdifferentiation of RECs.
a, Volcano plot of DEGs upon oestradiol induction of MYB74 expression. Upon the induction of MYB74, many photosynthesis-related genes are downregulated, whereas many genes related to cuticle formation are upregulated. NS, not significant. b, The Venn diagram illustrates that the majority of MYB74 target genes (2,176 of 2,834) are expressed in RECs. c, The Venn diagram shows the cell-state-specific expression of the MYB74 target genes that are expressed in RECs. d, Expression level of MYB74 in the individual single cells over pseudotime (top). The shaded band represents the 95% confidence interval. e, Signature score of 18 photosynthesis-related, 110 stress-response-related and 17 cutin and wax biosynthesis- and cuticle-development-related genes in the individual single cells (bottom). The shaded band represents the 95% confidence interval. f, Clustered dot plot showing shared state-specific cellular activities during REC transdifferentiation in putative MYB74 target genes, including the loss of photosynthetic competence and the acquisition of protective cuticle layer formation. g, Excess, ectopic cutin deposition in plants harbouring prUBQ10:XVE>>MYB74 after β-oestradiol treatment. The cutin deposition, which is shown in green, is visualized using auramine O. The cell walls are visualized using calcofluor white. Scale bar, 50 µm.
To provide further support for the role of MYB74 in transdifferentiation, we compared our scRNA-seq data with the 2,834 putative MYB74 target genes identified from a previous study using DNA affinity purification sequencing48. Notably, 2,176 of the MYB74 putative target genes were detected in the REC single-cell transcriptome (Fig. 3b and Supplementary Table 2). Of the 2,176 genes, 329 (15%) showed cell-state-specific expression (Fig. 3c), and 687 genes (32%) were upregulated or downregulated by MYB74 induction (Extended Data Fig. 7d and Supplementary Table 1). GO enrichment analysis of the 329 genes highlighted GO terms that overlap with those associated with scRNA-seq data at each state (Extended Data Fig. 7e and Fig. 1d). We further analysed MYB74 target genes that either show anticorrelation or correlation with the expression level of MYB74 in individual single cells. In agreement with Extended Data Fig. 6a,b, the expression level of MYB74 is low in the earlier stage, peaks in the middle stage and then appears to decrease in the later stage (Fig. 3d). The expression of photosynthesis-related genes anti-correlated with MYB74 expression over the course of transdifferentiation (Fig. 3e,f). By contrast, the expression of stress response genes showed strong correlation with the MYB74 expression level in state 2 and were notably downregulated in state 3 cells (Fig. 3e). Cutin biosynthesis-, wax biosynthesis- and cuticle development-related genes were highly upregulated, an event immediately following the upregulation of MYB74 (Fig. 3e,f). These results are similar to the expression patterns of photosynthesis- and cuticle-related genes in iMYB74 plants (Fig. 3a and Extended Data Fig. 7c). Furthermore, when we induced MYB74 expression using β-oestradiol, auramine O staining of the receptacle sections suggested that the ectopic cuticular material accumulated in the cell layers underneath the epidermal cells (Fig. 3g). No deposition of auramine O-stained cuticular materials was observed in the cells beneath the epidermis when MYB96 or SHN1 TFs, responsible for cuticular wax and cutin deposition, were overexpressed49,50. Taken together, our results imply that MYB74 mediates the conversion of nonepidermal RECs with photosynthetic competence into epidermal cells with a cuticle on the surface.
Transcriptional repressor MYB74-SRDX hinders fruit growth
Having linked MYB74 to REC transdifferentiation, we next investigated the phenotypes of a loss-of-function mutant for MYB74. We examined the expression pattern of two additional MYB genes in detail because there are other MYB genes whose expression pattern is similar to that of MYB74 and that were not included as cell-state-specific TFs owing to their low expression level (Extended Data Figs. 5b and 8a). MYB102 is the closest member to MYB74 in a phylogenetic tree51 and was therefore selected, while MYB2 was randomly selected. The expression pattern of both MYB102 and MYB2 in the AZ was very similar to that of MYB74 (Extended Data Fig. 8b,c). We found no detectable phenotypic change in the AZ of myb74 single-, myb74myb102 double- and myb74my102myb2 triple-knockout mutants (Extended Data Fig. 8d,e), suggesting that MYBs expressed in RECs are functionally redundant.
To circumvent this obstacle to genetic analysis, we used targeted transcriptional repression by expressing a chimeric MYB74 repressor under its native promoter, which leads to dominant repression of target genes52 (Extended Data Fig. 9a). Two independent lines of transgenic plants carrying prMYB74:MYB74-SRDX, lines 5 and 35, expressed high levels of the chimeric MYB74 repressor, with line 5 showing a higher level than line 35 (Extended Data Fig. 9b). As a first step in the analysis of prMYB74:MYB74-SRDX plants, we examined cuticle layer formation in the AZ of siliques at stage 17 (Fig. 4a). Compared with the wild-type (WT), toluidine blue stained the AZ of much older siliques of prMYB74:MYB74-SRDX plants, indicating notable impairment of cuticle formation. The deposition of lipid-rich cuticles was visualized by auramine O staining in the AZ of WT plants whereas the staining was compromised in the AZs of the two independent prMYB74:MYB74-SRDX lines (Fig. 4b). Scanning electron microscopy of the AZ surface also illustrated a clear difference between the WT and the prMYB74:MYB74-SRDX plants (Fig. 4b). In contrast to the WT, cells in the AZ of the prMYB74:MYB74-SRDX plants appeared to lack cuticular wax, consistent with the results of the toluidine blue permeability assay and comparable to the AZ of the transgenic plants in which the cutinase CDEF1 is driven by the ATML1 promoter (Fig. 4b and Extended Data Fig. 9c).
a, Impaired formation of a cuticle layer in the AZ of plants harbouring prMYB74:MYB74-SRDX. Cuticle formation and intactness are assessed by toluidine blue staining. Stage 17 siliques of the primary inflorescence at each flower position are shown. A representative result of three independent tests (>10 primary inflorescences for each test) is presented. Scale bars, 1 mm. b, SEM images show the altered cuticular wax deposition in the prMYB74:MYB74-SRDX plants as apparent using auramine O staining (left on each SEM image). Magnified images show representative auramine O-stained stamen AZ (shown in yellow). Scale bars, 25 µm. c, Impaired expression of GPAT4 in the RECs carrying prUBQ10>>MYB74-SRDX and prGPAT4:nls-GFP upon MYB74-SRDX induction via 1-day treatment with β-oestradiol. Stage 17 siliques from the primary inflorescence at each flower position were examined. Scale bar, 50 µm. d, Reduced expression of ATML1 in the RECs harbouring prMYB74:MYB74-SRDX and prATML1:nls-GFP. The cell walls are visualized using calcofluor white (b–d). Stage 17 siliques of the primary inflorescence at each flower position are shown. Scale bar, 50 µm. e,f, Reduced growth of receptacles (e) and fruits (f) of WT and prMYB74:MYB74-SRDX plants. Siliques at stage 17 from 27 independent plants were measured for each plant line (719 siliques, WT; 687 siliques, prMYB74:MYB74-SRDX line 5; 632 siliques, prMYB74:MYB74-SRDX line 35). The data indicate the mean (red centre line) ± s.d. (black lines) from three independent measurements. Data from each replicate are shown in different colours for each line. The bar graphs show distribution changes in receptacle width (e) and silique length in each line (f). Statistical significance was determined by one-way ANOVA followed by Fisher exact tests; *P < 0.0001.
GPAT4 encodes a glycerol-3-phosphate acyltransferase in the cutin biosynthesis pathway53 and is specifically expressed in the outermost RECs (Fig. 2d). GPAT4 is a potential target of the MYB74 TF48. We first verified that GPAT4 is indeed a target gene of MYB74 using the dual luciferase reporter assay54 (Extended Data Fig. 9d). Furthermore, we found that GPAT4 is co-expressed with MYB74 in the de novo specified epidermal cells (Extended Data Fig. 9e). The expression of MYB74-SRDX, driven by the UBQ10 promoter and upregulated by the β-oestradiol treatment, markedly diminished GPAT4 expression (Fig. 4c).
In addition to the cuticle layer survey, we further substantiated epidermal cell identity by analysing the expression of the epidermal cell marker ATML1, which is required for epidermal cell differentiation10. A recent study has suggested that ATML1 obtains a positional signal from very-long-chain fatty acid-containing ceramides, thereby contributing to epidermis differentiation in lateral root development55. ATML1 is first detected in the AZ from early stage 17 in WT, whereas the AZ of the prMYB74:MYB74-SRDX siliques barely exhibited expression of ATML1 (Fig. 4d). In addition, ATML1 expression was greatly reduced in transgenic plants in which the MYB74-SRDX repressor under the control of the UBQ10 promoter was induced by β-oestradiol, similar to the reduction in GPAT4 expression level (Extended Data Fig. 9f). This result shows that repressed MYB74 function hinders both epidermal identity and cuticle formation. It should be noted that MYB74 regulation of the epidermal marker ATML1 might be indirect because ATML1 was not identified as one of the MYB74 targets in the DAB-seq analysis. In addition, ATML1 expression is detected later in the state 3 cells following the expression of MYB74. Altogether, our findings strongly suggest that MYB74 plays a key role in the transdifferentiation of RECs into epidermal cells.
The receptacle undergoes notable radial growth following floral abscission (Fig. 2e), which involves the growth of the vasculature and pith. Given the defects in the epidermal cell features in the receptacle of the MYB74-SRDX plants, we investigated the consequences of the suppression of MYB74 function on receptacle growth after abscission. The MYB74-SRDX appears to inhibit radial growth, resulting in a reduced receptacle diameter (Fig. 4e) and a decrease in the distance from the pith centre to the AZ vasculature in the prMYB74:MYB74-SRDX lines (Extended Data Fig. 10a,b). Furthermore, we also found that the prMYB74:MYB74-SRDX plants had significantly shorter siliques (Fig. 4f and Extended Data Fig. 10d). At least 70–80% of the receptacles are covered with new surface cells after petals, stamens and sepals are shed (Fig. 4b–d and Extended Data Figs. 6a and 9c). As a result, a significant number of the new surface cells of the receptacles acquire epidermal identity. As epidermal cells contribute to the differentiation of internal tissues and plant growth, these results imply that de novo specified epidermal cells in the AZ also play a role in promoting plant growth. These findings collectively indicate that the repression of MYB74 affects both surface integrity and receptacle growth, thereby limiting fruit growth.
Discussion
In this work, we establish why and how RECs transdifferentiate into epidermal cells following abscission to reconstitute the protective cuticle layer. Using scRNA-seq, we characterize the gene expression changes that drive REC transdifferentiation through three distinct states to acquire epidermal cell identity. We show that MYB74 is a key transcriptional regulator that orchestrates the conversion of photosynthetic chloroplasts to nonphotosynthetic plastids, upregulation of stress response genes and deposition of the cuticle throughout this transdifferentiation process.
Our results indicate that hindering de novo specification of epidermal cells affects receptacle growth. Receptacles are located at the nodal region between the pedicel and the fruit, transporting water and nutrients to fruits and developing seeds56. In contrast to the established wound healing pathway that uses suberin and lignin to form an alternative protective layer after epidermal cell loss12,13,57, we show that epidermal transdifferentiation supports subsequent fruit growth at the abscission site (Fig. 4)58. Our findings suggest that the impaired de novo specification of epidermal cells leads to alterations in cellular structures in the receptacle, resulting in restricted fruit growth.
High levels of lignin accumulation can hinder plant growth59,60. It is interesting that MYB74 targets several MYB genes involved in the suppression of lignification, which are upregulated in state 2 RECs and the iMYB74 plants (Extended Data Fig. 10c). MYB4, MYB7 and MYB32 belong to the same subclade of the MYB phylogenetic tree and have been implicated in the repression of lignification61,62,63. MYB3 and MYB6 function as transcriptional repressors that reduce lignin biosynthesis64,65. Thus, it is conceivable that MYB74 regulates cuticle accumulation and, potentially, lignification during the transdifferentiation of RECs in favour of epidermal transdifferentiation and cuticle formation. Overall, our results imply that the newly exposed REC surface undergoes de novo epidermis formation not only to prevent pathogen invasion and tissue dehydration but also to assure fruit growth—in contrast to wound healing processes that could limit subsequent receptacle and fruit growth.
Importantly, our findings could inform new strategies to tune the balance between lignification and cuticle formation in crop plants to promote enhanced fruit growth. From a fundamental research perspective, the de novo specification of epidermal cells from RECs provides a new model system in which to study the interplay between the epidermis and its neighbouring cell types, overcoming the genetic lethality experienced by mutants with defective epidermal development. Overall, our work uncovers the genetic underpinnings of a critical plant developmental switch that allows plants to protect a vulnerable new surface exposed by abscission while facilitating continued tissue and fruit growth.
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown in soil in a growth chamber under the following growing conditions: 22 ± 2 °C with 110 µmol m−2 s−1 of light intensity under a 16-h photoperiod. For all experiments, seed stratification was performed at 4 °C for 2–5 days in the dark. All mutants used in this study were obtained from the Arabidopsis Biological Resource Center and include the following: ida-12 (SALK_133209), hae-1 (SALK_105975C), hsl2-1 (SALK_057117), myb74 (SM3.16165), myb2 (SALK_045455) and myb102 (SM3.41645).
Plasmid construction and generation of transgenic plants
Constructs were generated using a Gateway cloning (Invitrogen) protocol. For a complete list of constructs and relevant primer information, see Supplementary Table 3. Constructs were introduced into plants using the floral dip method, and transformed plants were selected on a medium containing antibiotics as previously described66. Briefly, Arabidopsis flower buds were dipped in a solution of 5% (w/v) sucrose, 0.05% (v/v) Silwet L-77 and Agrobacterium tumefaciens for 2–10 s, and were then placed in the dark overnight at high humidity. To examine the effect of MYB74-SRDX on marker gene expression, the prATML1:nls-GFP-GUS and prGPAT4:nls-GFP-GUS constructs were introduced into T2 transgenic plants carrying prMYB74:MYB74-SRDX and prUBQ10:XVE>>MYB74-SRDX, respectively. To examine the co-expression of marker genes, elite lines of prGPAT4 nls-mCherry and prHAE:nls-mCherry were crossed with prMYB74:nls-GFP-GUS marker lines, and F1 plants were analysed.
Preparation of protoplasts and fluorescence-activated cell sorting
Protoplasts of RECs of the receptacle were isolated from the AZ of Arabidopsis plants carrying the prEXL1:nls-GFP construct as previously described20 with minor modifications. Briefly, receptacle materials were collected at floral stages 15–17 as previously defined24,25 from approximately 520 plants. The receptacle materials were chopped in distilled water using a Waring WPB05 blender at high speed, twice for 30 s each time, and collected on Kimwipes (KIMTECH) to remove excess water. The chopped materials were transferred to an enzyme solution (0.5% macerozyme R-10, 1.2% cellulose R-10, 400 mM mannitol, 10 mM CaCl2, 4 mM 2-(N-morpholino)ethanesulfonic acid (MES, pH 5.7), 20 mM KCl, 2.5 mM β-mercaptoethanol, 0.1% bovine serum albumin, 40 U ml−1 RNase inhibitor and 10 μg ml−1 actinomycin D) and incubated in the dark at room temperature with gentle agitation (80 rpm) for 4 h. To facilitate infiltration of the enzyme solution, a vacuum (100 mm Hg) was applied for 30 min. Subsequently, isolated protoplasts were diluted with the same volume of ice-cold Ca2+-free washing and incubation solution (500 mM mannitol, 20 mM KCl and 4 mM MES, pH 5.7), filtered through a 40-μm cell strainer (BD Falcon), and the resulting solution was concentrated by centrifugation at 500 g for 5 min. The pelleted protoplasts were then resuspended in 5 ml washing and incubation solution in a 15-ml tube (BD Falcon), and GFP-positive protoplasts were collected using a FACSAris III cell sorter (BD Bioscience) with a 100-μm nozzle and a system pressure of <20 psi (ref. 67). Approximately 100,000 GFP-positive cells were collected into 1.5-ml tubes containing ice-cold PBS solution, which were subsequently used for scRNA-seq.
scRNA-seq
scRNA-seq libraries were generated using the Chromium Single Cell 3′ Library and Gel Bead Kit v2 (PN-120267), the Chromium Single Cell A Chip kit (PN-1000009) and the Chromium i7 Multiplex Kit (PN-120262), all from 10X Genomics. Library preparation was performed according to the manufacturer’s instructions outlined in the CG00052 Chromium Single Cell 3′ Reagent Kits v2 User Guide Rev B. First, protoplasts were counted, and their concentration was adjusted to 5 × 105–2 × 106 cells ml−1. Approximately 13,000 protoplasts were mixed with the reverse transcription master mix to capture 8,000 cells, which were then loaded onto a Single Cell A Chip (PN-1000009). Next, the Single Cell 3′ Gel Bead suspension and Partitioning Oil were loaded into the designated wells. After loading the reagents, the Single Cell A Chip was placed on the Chromium Controller to generate Gel Bead-In-Emulsions (GEMs). The GEMs were collected in tubes, and reverse transcription (GEM-RT) was performed using a thermocycler. During the GEM-RT incubation, the read 1 primer sequence (R1) was incorporated into the cDNA for Illumina library sequencing. Following cleanup of the GEM-RT reaction, the cDNA was amplified with a thermocycler. The amplified cDNA was then analysed qualitatively using an Agilent Bioanalyzer High Sensitivity chip (Agilent). Enzymatic fragmentation and size selection were conducted to optimize the cDNA amplicon size. The subsequent library preparation steps, including end repair, A-tailing, adaptor ligation and sample index PCR, were carried out to incorporate the P5, P7, the read 2 primer sequence (R2) and sample indices. Finally, the libraries were sequenced on an Illumina HiSeq 4000 platform using 2 × 100 bp paired-end reads with a target mean of 50,000 read pairs per cell.
scRNA-seq data preprocessing and quality control
After sequencing, raw reads were mapped to the Arabidopsis reference genome using Cell Ranger version 2.1.1 and the Ensembl GTF file (TAIR10.40). Empty droplets were removed using the DropletUtils version 1.14.2 R package68 with a false discovery rate (FDR) < 0.05. We classified cells as low quality if they had <1,000 unique molecular identifiers (UMIs) or if >10% of UMIs mapped to mitochondrial and chloroplast genes; low-quality cells were excluded from downstream analyses. Raw UMI counts were then normalized by cell-specific size factors estimated using the scran version 1.22.1 R package69 with all options set to the default and log2 transformed using a pseudo-count of 1. Highly variable genes were identified using the modelGeneVar function of scran with FDR < 0.1 and biological variability >0.01. Following the removal of non-RECs (that is, secession cells, guard cells and vasculature cells), principal component analysis of the scaled expression matrix of highly variable genes was performed using the RunPCA function of the Seurat R package for clustering and visualization of the RECs. All RECs were grouped into five clusters and visualized in a two-dimensional Uniform Manifold Approximation and Projection (UMAP) plot using the FindClusters and RunUMAP functions of the Seurat package with the first 200 principal components (PCs). These five clusters were assigned to one of three states according to their correlations.
Signature scores
Gene set signature scores were computed using the AddModuleScore function of the Seurat version 4.1.1 R package70. For non-RECs, signature scores were calculated using canonical marker genes. The state-specific MYB74 target gene scores were estimated from potential MYB74 target genes associated with photosynthesis (GO:0015979), stress response (GO:0006950) and cuticle synthesis (GO:0010143, GO:0160062, GO:0042335 and GO:0010025).
GO enrichment analysis of scRNA-seq data
For each transdifferentiation state, marker genes were identified using the FindAllMarkers function of the Seurat package with default options. Genes with adjusted P < 0.05 were considered to be state-specific markers. The level of GO term enrichment for each state was calculated for the marker genes using the gProfiler2 version 0.2.1 R package71 with all options set to the defaults.
Trajectory inference
To infer the state transition of transdifferentiation, trajectory inference was performed using the Palantir version 1.0.1 Python package72. Diffusion components were computed using the run_diffusion_maps function of Palantir for the first 10 PCs. Next, a k-nearest neighbour graph was constructed using the first 10 diffusion components with k = 50. To visualize these results, a two-dimensional ForceAtlas2 (FA) plot was computed using the tl.draw_graph function of the scanpy version 1.9.1 Python package73 with all options set to default values. Pseudotimes were inferred using the core.run palantir function of Palantir with the option num_waypoints = 500. Finally, the starting cell was selected based on the highest photosynthesis signature score.
RNA velocity
To support the inferred trajectories, RNA velocities were estimated using the Python package scVelo (version 0.2.3)74. The spliced and unspliced count matrices were generated using the Python package velocyto (version 0.17.17)75. After genes with low expression levels were filtered out, all matrices were normalized and transformed to natural logarithm values using the pp.filter_and_normalize function with the options n_top_genes = 500 and min_shared_counts = 5. For each cell, the moments for velocity estimation were computed using the pp.moments function using the options n_neighbors = 30 and n_pcs = 15. RNA velocities were estimated using the tl.velocity function with all options set to default parameters. The velocity graphs were then computed from the estimated velocities using the tl.velocity_graph function, again using the default options. The RNA velocity results were visualized on a two-dimensional FA plot.
Cell-type label transfer
Cell-type label transfer was conducted using the Seurat package to predict the identities of RECs. To facilitate label transfer between different datasets, we identified anchors, which are mutual nearest neighbours in the reference and query datasets. These anchors serve as reference points to align the datasets and facilitate the transfer of cell-type labels. The anchors between the reference scRNA-seq datasets27 and the RECs were identified using canonical correlation analysis through the FindTransferAnchors function with the parameter reduction = “cca”, while keeping the default settings for the other parameters. Finally, cell-type labels from the reference dataset were transferred to the RECs using the first five canonical correlation vectors.
Inducible expression of MYB74 (iMYB74) or MYB74-SRDX (iMYBY74-SRDX) in Arabidopsis
The coding sequences of MYB74 and MYB74-SRDX were cloned into the oestradiol-inducible binary vector pMDC7 (ref. 47). Next, the pMDC7-MYB74 and pMDC7-MYB74-SRDX constructs were introduced into Arabidopsis plants using Agrobacterium-mediated floral dipping. To evaluate the effect of iMYB74 on the transcriptome, receptacles at floral stages 15–17 were collected from 5- to 6-week-old iMYB74 plants. Receptacles were then placed in a one-half MS medium supplemented with 10 μM β-oestradiol and incubated for 10 h under gentle agitation at 70 rpm. After incubation, total RNA was extracted from the samples using the RNeasy Mini Kit (Qiagen). The total RNA concentration was then quantified using a Nanodrop spectrophotometer (Thermo), and RNA quality was assessed using a Bioanalyzer equipped with an RNA Pico chip (Agilent Technologies). To investigate the effects of iMYB74 or iMYB74-SRDX on REC transdifferentiation, main inflorescence stems were collected from 5-week-old iMYB74 or iMYB74-SRDX plants and placed in 2-ml tubes containing one-half MS medium supplemented with or without 10 μM β-oestradiol. After 1 to 5 days of this treatment, receptacles were prepared for examination by confocal microscopy.
Bulk RNA-seq data processing and analysis
Paired-end sequencing reads were generated using an Illumina NovaSeq sequencing platform. Before downstream analyses, Trimmomatic version 0.38 was used to remove adaptor sequences and trim low-quality bases. The cleaned reads were then aligned to the A. thaliana genome assembly (TAIR10.1) using HISAT version 2.1.0 (ref. 76) based on the HISAT and Bowtie2 implementations. The reference genome sequence and gene annotation data were downloaded from the NCBI Genome Assembly and NCBI RefSeq databases, respectively. Aligned data (in the SAM file format) were then sorted and indexed using SAMtools version 1.9. After alignment, transcripts were assembled and quantified using StringTie version 2.1.3b77,78. Finally, gene- and transcript-level quantifications were performed to calculate raw read counts, fragments per kilobase of transcript per million mapped reads values and transcripts per million values.
Differential gene expression (DEG) analysis
Statistical analysis of differential gene expression was performed using DESeq2 (ref. 79). Quality control was performed by selecting genes with nonzero counts in all samples, and multidimensional scaling plots were generated to observe the expression similarity between samples. A filtered dataset was applied with Relative Log Expression normalization to correct for variation in library size among samples. The statistical significance of differential gene expression was then determined using a DESeq2 nbinom Wald Test79, and fold change and P values were extracted from Wald test results. All P values were adjusted using the Benjamini–Hochberg algorithm to control the FDR. Genes identified as significantly differentially expressed were those showing |fold change| ≥ 2 and raw P < 0.05. Hierarchical clustering on significant genes was then performed using rlog-transformed values using the following parameters: distance metric = Euclidean distance and linkage method = complete. Next, gene-enrichment and functional annotation procedures for significant differentially expressed genes (DEGs) were carried out using gProfiler80 against the GO and Kyoto Encyclopedia of Genes and Genomes pathway (https://www.genome.jp/kegg/) databases. Finally, adjusted P values reported from gProfiler results were derived using a one-sided hypergeometric test and statistical tests were corrected using the Benjamini–Hochberg method.
Data analysis and DEG visualization were then conducted using R version 3.6.1 (www.r-project.org). Biological process GO enrichment analysis was performed using GO enrichment analysis tools listed at https://geneotology.org (ref. 81). ShinyGO version 0.77 was also used for biological process GO and Kyoto Encyclopedia of Genes and Genomes analysis and to generate graphical representations for all data82.
Scanning electron microscopy and histology
Scanning electron microscopy (SEM) analysis of floral organ AZs was performed as described previously20. Sepals and petals were first removed from flowers before fixation. Following critical-point drying using an EM CPD300 dryer (Leica), tissue samples were mounted onto steel stubs, coated with osmium then observed under a Hitachi S-4800 SEM. Histological analysis of AZs was performed using samples embedded in Technovit 7100 resin (Heraeus Kulzer) using a previously described method20. Finally, embedded samples were cut into 2-μm sections using an RM2255 microtome (Leica) with TC65 tungsten carbide disposable blades (Leica), stained with 0.1% toluidine blue and observed under a Zeiss Axiovert 40 inverted microscope. The radius of the receptacles from the AZ vasculature to the pith centre was measured in the sections on the acquired photographs, using AxioVision software.
Gene expression analysis using promoter-GUS reporters
Promoter-driven β-glucuronidase (GUS) activity was examined as described previously20. Briefly, flowers at various stages of development were collected and incubated in GUS solution for 1–24 h at 37 °C. Subsequent steps involved chemical fixation, gradual dehydration in an ethanol series and finally embedding in Technovit 3040 solution. Embedded samples were then sectioned and imaged as described above. The AxioVision software was used to capture photographs.
Tissue clearing and confocal laser scanning microscopy
The expression of marker genes was analysed using nls-GFP or nls-mCherry reporters driven by native promoters. To assess expression, the flowers of marker lines were first longitudinally or transversely sectioned, fixed in 4% PFA, cleared in ClearSee solution29 and then counterstained with cell wall dye (0.01% calcofluor white). After rinsing, the cleared materials were mounted on slides with ClearSee solution and were observed under a Zeiss LSM710 confocal scanning microscope. To visualize cuticle deposition, 0.01% Auramine O was used as previously described29,83.
Light-sheet microscopy
Light-sheet images were then taken from flowers of marker lines. Flowers were first fixed with 4% PFA, cleared in ClearSee solution for two weeks, then counterstained with 0.1% calcofluor white29. Next, cleared flowers were rinsed in ClearSee solution and embedded in 1% low-melting-temperature agarose in glass capillaries before imaging using a Lightsheet Z.1 microscope (Carl Zeiss).
Permeability assay of the AZ
The permeability of the AZ was examined by assessing the penetration of 0.03% toluidine blue, as previously described20. Briefly, flowers at stages 15–17 were harvested from wild-type and transgenic plants and simultaneously incubated in a 0.03% toluidine blue solution for 2–3 min. Stained flowers were quickly washed in water and photographed under a Zeiss V12 stereomicroscope equipped with motorized zoom and an Axiocam 512 colour camera. This experiment was repeated three times. ImageJ was used to measure the stained area.
Measuring receptacle width and silique length
The physical dimensions of the receptacle and silique were determined. To do so, siliques were chosen at floral stage 17 from the main inflorescence stems of both wild-type and transgenic plants. The siliques were lined up with two median nectaries facing up using double-sided adhesive tape then photographed under a Zeiss V12 stereomicroscope equipped with an Axiocam 512 colour camera. Receptacle width and silique length were then measured from the acquired photographs using AxioVision software. A total of 27 main inflorescence stems were analysed per genotype.
Dual luciferase assay and transient expression in Nicotiana benthamiana
The GPAT4 promoter (~2 kb) sequence and the MYB74 coding sequence were cloned in the pGreenII 0800-LUC Gateway vector54 and pEarlyGate100 (pEG100) vector, respectively. The Multi Cloning Site from TSK108 inserted into the pEG100 vector was used as an empty vector control. Agrobacterium (GV3101) cultures transformed with each pGreenII 0800-GPAT4p-LUC (prGPAT4:LUC), pr35S:MYB74 and pr35S:MCS (multi-cloning site from TSK108) construct were resuspended in an infiltration buffer (10 mM MES (pH 5.8), 10 mM MgCl2, 150 μM acetosyringone). Five-week-old N. benthamiana leaves were then co-infiltrated with P19 and a 1:9 ratio of GPAT4p-LUC and the TF set as used in the previously described experiment54. Three days after infiltration, leaf discs were snap-frozen in liquid N2, ground to a powder using a TissueLyser and then diluted in 100 μl passive lysis buffer (Promega). Next, extracts were centrifuged at 8,000 g for 1 min, after which 65-µl aliquots of the supernatant were examined using the Dual-Glo Luciferase Assay System (Promega)84 with all procedures following the manufacturer’s instructions. Luminescence was detected using an Infinite M200 PRO multimode plate reader (Tecan).
RT-qPCR analysis
Total RNA was extracted from receptacle samples using the RNeasy Micro Kit (QIAGEN) with all procedures performed according to the manufacturer’s instructions. cDNA was then synthesized using 1 µg of total RNA and qScript cDNA SuperMix (QUANTABIO) as per the manufacturer’s instructions. Relative expression was then measured in triplicate by RT-qPCR using a LightCycler480 II system (Roche) and a SYBR Green I Master kit (Roche) with all procedures following the manufacturer’s instructions. Data were analysed using qBase+ (Biogazelle). Expression levels were normalized to those of UBQ10 and eIF4A. All primers used for RT-qPCR analysis are listed in Supplementary Table 3.
Statistical analysis and reproducibility
All quantitative data were analysed using Origin2021 (Origin Lab, https://www.originlab.com/) or GraphPad Prism. Plots were created using Origin2021, GraphPad or ggplot2 (v. 3.4.3). Statistical significance was determined using two-sided Student’s t-tests or one-way ANOVA with Tukey’s honestly significant difference method, with a significance threshold set at P < 0.05. Individual P values are provided in the results. Bar graphs display the mean ± s.d., while box plots show the 25th to 75th percentiles (box limits), the full data range (whiskers) and the median (centre line). Open or filled dots represent individual samples in both bar and box plots. Experiments were independently repeated at least three times, and representative images are shown unless specified otherwise in the figure legends, which include details on sample size and the number of biological replicates (n). For the analyses of transgenic plants, two to three representative lines were selected from 16 to 32 independently transformed plants after confirming consistent expression patterns of the introduced reporters or phenotypic traits. All analyses were conducted on the T2 or T3 generations of the representative transgenic lines.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data generated in this study are included in the article and Supplementary Information (Supplementary Tables 1–3). Single-cell RNA-seq data (accession number PRJNA1013602) and bulk RNA-seq data (accession number PRJNA1032145) generated in this study are available at the Sequence Read Archive database. Requests for materials and correspondence should be addressed to corresponding author J.M.K.
Code availability
The code used to analyse the scRNA-seq data in this study is available via GitHub at https://github.com/CB-postech/scRNA-seq_prEXL1_REC.
References
Javelle, M., Vernoud, V., Rogowsky, P. M. & Ingram, G. C. Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39 (2011).
Asaoka, M. et al. Stem integrity in Arabidopsis thaliana requires a load-bearing epidermis. Development 148, dev198028 (2021).
Yeats, T. H. & Rose, J. K. The formation and function of plant cuticles. Plant Physiol. 163, 5–20 (2013).
Kutschera, U. & Niklas, K. J. The epidermal-growth-control theory of stem elongation: an old and a new perspective. J. Plant Physiol. 164, 1395–1409 (2007).
Savaldi-Goldstein, S., Peto, C. & Chory, J. The epidermis both drives and restricts plant shoot growth. Nature 446, 199–202 (2007).
Nobusawa, T. et al. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 11, e1001531 (2013).
Bidhendi, A. J., Altartouri, B., Gosselin, F. P. & Geitmann, A. Mechanical stress initiates and sustains the morphogenesis of wavy leaf epidermal cells. Cell Rep. 28, 1237–1250 (2019).
Malivert, A., Hamant, O. & Ingram, G. The contribution of mechanosensing to epidermal cell fate specification. Curr. Opin. Genet. Dev. 51, 52–58 (2018).
Hamant, O. et al. Developmental patterning by mechanical signals in Arabidopsis. Science 322, 1650–1655 (2008).
Takada, S. & Iida, H. Specification of epidermal cell fate in plant shoots. Front. Plant Sci. 5, 49 (2014).
Kurdyukov, S. et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18, 321–339 (2006).
Wei, X., Guan, W., Yang, Y., Shao, Y. & Mao, L. Methyl jasmonate promotes wound healing by activation of phenylpropanoid metabolism in harvested kiwifruit. Postharvest Biol. Technol. 175, 111472 (2021).
Han, Y., Yang, R., Wang, Q., Wang, B. & Prusky, D. Sodium silicate promotes wound healing by inducing the deposition of suberin polyphenolic and lignin in potato tubers. Front. Plant Sci. 13, 942022 (2022).
Lulai, E. C. & Neubauer, J. D. Wound-induced suberization genes are differentially expressed, spatially and temporally, during closing layer and wound periderm formation. Postharvest Biol. Technol. 90, 24–33 (2014).
Nawrath, C. et al. Apoplastic diffusion barriers in Arabidopsis. Arabidopsis Book 2013, e0167 (2013).
Estornell, L. H., Agusti, J., Merelo, P., Talon, M. & Tadeo, F. R. Elucidating mechanisms underlying organ abscission. Plant Sci. 199, 48–60 (2013).
Sexton, R. & Roberts, J. A. Cell biology of abscission. Annu. Rev. Plant Physiol. 33, 133–162 (1982).
Poovaiah, B. W. Formation of callose and lignin during leaf abscission. Am. J. Bot. 61, 829–834 (1974).
Niederhuth, C. E., Cho, S. K., Seitz, K. & Walker, J. C. Letting go is never easy: abscission and receptor-like protein kinases. J. Integr. Plant Biol. 55, 1251–1263 (2013).
Lee, Y. et al. A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173, 1468–1480 (2018).
Ingram, G. & Nawrath, C. The roles of the cuticle in plant development: organ adhesions and beyond. J. Exp. Bot. 68, 5307–5321 (2017).
Hawkins, S. & Boudet, A. Wound-induced lignin and suberin deposition in a woody angiosperm (Eucalyptus gunnii Hook): histochemistry of early changes in young plants. Protoplasma 191, 96–104 (1996).
Xie, P. D. et al. Chitooligosaccharide accelerated wound healing in potato tubers by promoting the deposition of suberin polyphenols and lignin at wounds. Plant Physiol. Biochem. 199, 107714 (2023).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Cai, S. & Lashbrook, C. C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146, 1305–1321 (2008).
Shahan, R. et al. A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Dev. Cell 57, 543–560 (2022).
Lopez-Anido, C. B. et al. Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev. Cell 56, 1043–1055 (2021).
Waters, M. T. et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128 (2009).
Ursache, R., Andersen, T. G., Marhavy, P. & Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018).
Chang, H. C., Tsai, M. C., Wu, S. S. & Chang, I. F. Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana. Bot. Stud. 60, 16 (2019).
Wei, N. et al. Genome-wide identification of ERF transcription factor family and functional analysis of the drought stress-responsive genes in Melilotus albus. Int. J. Mol. Sci. 23, 12023 (2022).
Alvarez-Buylla, E. R. et al. Flower development. Arabidopsis Book 2010, e0127 (2010).
Ramsay, N. A. & Glover, B. J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70 (2005).
Lai, L. B. et al. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17, 2754–2767 (2005).
Lee, S. B., Kim, H. U. & Suh, M. C. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 57, 2300–2311 (2016).
Kosma, D. K. et al. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. 80, 216–229 (2014).
Cominelli, E., Sala, T., Calvi, D., Gusmaroli, G. & Tonelli, C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53, 53–64 (2008).
Cominelli, E., Galbiati, M. & Tonelli, C. Integration of water stress response: cell expansion and cuticle deposition in Arabidopsis thaliana. Plant Signal. Behav. 3, 556–557 (2008).
Xu, H. M. et al. Transcriptional networks regulating suberin and lignin in endodermis link development and ABA response. Plant Physiol. 190, 1165–1181 (2022).
Cesarino, I. With a little help from MYB friends: transcriptional network controlling root suberization and lignification. Plant Physiol. 190, 1077–1079 (2022).
Liljegren, S. J. Organ abscission: exit strategies require signals and moving traffic. Curr. Opin. Plant Biol. 15, 670–676 (2012).
Cho, S. K. et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 105, 15629–15634 (2008).
Jinn, T. L., Stone, J. M. & Walker, J. C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117 (2000).
Stenvik, G. E. et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817 (2008).
Ortiz-Garcia, P. et al. The indole-3-acetamide-induced Arabidopsis transcription factor MYB74 decreases plant growth and contributes to the control of osmotic stress responses. Front. Plant Sci. 13, 928386 (2022).
Xu, R. et al. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 66, 5997–6008 (2015).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
O’Malley, R. C. et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016).
Seo, P. J. et al. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152 (2011).
Kannangara, R. et al. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294 (2007).
Dubos, C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010).
Hiratsu, K., Matsui, K., Koyama, T. & Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 (2003).
Yang, W. et al. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol. 160, 638–652 (2012).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Nagata, K., Ishikawa, T., Kawai-Yamada, M., Takahashi, T. & Abe, M. Ceramides mediate positional signals in Arabidopsis thaliana protoderm differentiation. Development 148, dev194969 (2021).
Guo, X., Xiao, X., Wang, G. & Gao, R. Vascular anatomy of kiwi fruit and its implications for the origin of carpels. Front. Plant Sci. 4, 391 (2013).
Paul-Victor, C. et al. Effect of mechanical damage and wound healing on the viscoelastic properties of stems of flax cultivars (Linum usitatissimum L. cv. Eden and cv. Drakkar). PLoS ONE 12, e0185958 (2017).
Ripoll, J. J. et al. Growth dynamics of the fruit is mediated by cell expansion. Proc. Natl Acad. Sci. USA 116, 25333–25342 (2019).
Zhou, J., Lee, C., Zhong, R. & Ye, Z. H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266 (2009).
Neves, G. Y. S., Marchiosi, R., Ferrarese, M. L. L., Siqueira-Soares, R. C. & Ferrarese, O. Root growth inhibition and lignification induced by salt stress in soybean. J. Agron. Crop Sci. 196, 467–473 (2010).
Jin, H. et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161 (2000).
Zhao, Q. & Dixon, R. A. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233 (2011).
Xiao, R. X., Zhang, C., Guo, X. R., Li, H. & Lu, H. MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 22, 3560 (2021).
Wang, L. et al. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751 (2019).
Kim, D. et al. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 41, 1549–1560 (2022).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Birnbaum, K. et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Lun, A. T., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J. & Peterson, H. gprofiler2—an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 9, ELIXIR-709 (2020).
Setty, M. et al. Author correction: Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 1237 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Thomas, P. D. et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22 (2022).
Ge, S. X., Jung, D. M. & Yao, R. A. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020).
Lequeu, J., Fauconnier, M. L., Chammai, A., Bronner, R. & Blee, E. Formation of plant cuticle: evidence for the occurrence of the peroxygenase pathway. Plant J. 36, 155–164 (2003).
Seo, H. et al. Intragenic suppressors unravel the role of the SCREAM ACT-like domain for bHLH partner selectivity in stomatal development. Proc. Natl Acad. Sci. USA 119, e2117774119 (2022).
Acknowledgements
We thank Y. Lee (Seoul National University) for her initial contribution and advice. We also thank J. Jeon, J. Choi and M. Kim for their technical support. We are grateful to W.T. Kim (Yonsei University), P.O. Lim (DGIST), Z.-M. Pei (Duke University) and Life Science Editors for their critical reading of the paper. This work was supported by grants from Samsung Science and Technology Foundation (SSTF-BA2101-10) and the National Research Foundation (grant number: 2022R1A2C3007309) and in part by a grant from DGIST (22-CoE-BT-04) to J.M.K.
Author information
Authors and Affiliations
Contributions
J.M.K. conceived the study, and J.M.K., J.K.K., J.-U.H., S.K. and X.W. designed the experiments. C.W.L., E.L., D.G.C. and J.K.K. analysed the scRNA-seq. S.K., J.-U.H., S.-K.H. and J.K.K. analysed the bulk RNA-seq data. X.W., S.K., J.-U.H., Y.H.C., S.-K.H., T.-H.Y., S.L., H.S., S.H.J., J.L., H.L. and H.C. performed the experiments. X.W., S.K., C.W.L., J.-U.H., J.K.K. and J.M.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Nobutoshi Yamaguchi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single-cell RNA-seq analysis of residuum cells.
a, Workflow for single-cell RNA-sequencing (scRNA-seq) analysis of prEXL1:nls-GFP-expressing residuum cells (RECs). Receptacle tissues were collected, which are indicated by dotted yellow rectangles. Prior to protoplasting, tissues were observed under bright-field and GFP fluorescence to confirm GFP expression. After protoplasting, approximately 100,000 GFP-positive cells were isolated via fluorescence-activated cell sorting. These cells were then subjected to droplet-based scRNA-seq. Following raw data filtering, we obtained high-quality data for further analysis. b, A Uniform Manifold Approximation and Projection (UMAP) projection before the removal of non-RECs. After performing the dimensional reduction on 3,489 individual transcriptomes from AZ cells, we identified 23 cell clusters, each represented by a different color. Cell clusters corresponding to RECs are labeled with bold red numbers, whereas non-RECs are labeled with black numbers. c, UMAP plots showing the signature scores of non-REC cells, which are calculated based on the expression levels of cell type-specific marker genes. d, FACS gating strategy for GFP-positive REC protoplasts. Debris and doublets were excluded based on the forward scatter (FSC) and side scatter (SSC) properties. This is achieved by comparing FSC area (FSC-A) vs. SSC area (SSC-A), FSC width (FSC-W) vs. FSC height (FSC-H), and SSC width (SSC-W) vs. SSC height (SSC-H) sequentially. GFP-positive protoplasts were identified based on fluorescence intensity in the FITC channel, using FITC-A vs. FSC-A to differentiate cell populations. The final gated GFP-positive protoplasts were then isolated for downstream analysis.
Extended Data Fig. 2 scRNA-seq analysis identifies three distinct cell states during REC transdifferentiation.
a, UMAP projection yields five major clusters after removing non-RECs, showing the distribution of 2,823 RECs. Each cluster is color-coded. b–d, Violin plots displaying cell quality control statistics for the 2,823 RECs (n = 1): (b) the total number of unique molecular identifiers (UMIs), (c) the number of detected genes, and (d) the percentage of mitochondrial and chloroplast mRNA content per cell. The box plots display the median and interquartile range (25th-75th percentiles). The whiskers extend to 1.5 times the interquartile range. e, Pearson correlation analysis reveals three distinct developmental states among the five cell clusters. The colored bar indicates the scaled correlation. The color code for the cell clusters is consistent with that used in (a). f, UMAP projection shows the distribution of 2,823 cells across three developmental states. g, Heatmap illustrating gene expression correlations between cell states, calculated using Spearman rank correlation analysis. Cells are ordered along pseudotime. Within each pseudotime bin, cell expression profiles were averaged before calculating correlation coefficients. The color-coded bar at the top represents the relative proportion of developmental states within each pseudotime bin. h, Violin plots showing intercellular Spearman rank correlation coefficients for each developmental state from the 2,823 RECs (n = 1). Statistical significance was determined using unpaired two-sided Student’s t-tests (****p < 0.0001; state 1 vs 2: p < 2e-16, state 1 vs 3: p < 2e-16, state 2 vs 3: p < 2e-16). The box plots display the median and interquartile range (25th-75th percentiles) and the whiskers extend to 1.5 times the interquartile range. i, The heatmap shows the correlation between RECs in different developmental states and four inferred cell types after transfer analysis using Arabidopsis leaf scRNA-seq datasets27. Values were calculated using Pearson correlation coefficient analysis. The same color code was used for developmental states, as in panels e–i.
Extended Data Fig. 3 Expression profiles of cell state-specific genes during REC transdifferentiation.
a, Venn diagram illustrates the number of genes preferentially expressed in each developmental state. b, Heatmap displays the expression dynamics of cell state-specific genes over pseudotime. The mean gene expression values (that is, z-scores) are shown within each pseudotime bin. The color-coded bar at the top represents the relative proportion of developmental states within each pseudotime bin. The same cell state color code was used for all main and extended data figures.
Extended Data Fig. 4 Expression pattern of marker genes for cell states 2 and 3.
a, Expression of prABI5:nls-GFP (top) and prERF1A:nls-GFP (bottom) in receptacle AZs before and after abscission. Z-projected surface scan images show the spatiotemporal expression patterns of ABI5 and ERF1A in AZs over successive stages, thereby confirming the results of our scRNA-seq analysis. Scale bars, 100 µm. b, Expression patterns of state 3-specific genes (that is, BDG1, GPAT4, KCS20, LACS2, LTPG2, and WRI4) in AZs from S15 to S17m, as examined using promoter:nls-GFP reporters. Scale bars, 100 µm. c, Section images show the expression of KCS20, WRI4, LACS2, and LTPG2 only in the outermost layer of RECs after abscission (S17m), indicating that only one layer of RECs differentiates into epidermal cells. Scale bar, 50 µm.
Extended Data Fig. 5 Schematic diagram illustrating the progression of REC transdifferentiation and expression profiles of the cell state-specific transcription factors.
a, The schematic drawing illustrates which stages of fruit development correspond to the cellular states 1, 2, and 3. At flower development stage 15 (S15), sepals, petals, and stamens wilt and undergo abscission. Stages S15e (early) and S15l (late) indicate the flower stages immediately following S14 and preceding S16, respectively. At S16, the fruit is approximately twice the length of the petals. S17 includes stages beyond S16, with specific substages defined as follows: S17e (early) corresponds to fruit elongation, S17m (middle) to fruit lateral growth, and S17l (late) to the beginning of yellowing at the fruit tip. These stages also align with phases of embryo development: S17e relates to the globular to early heart stages, S17m corresponds to the late heart to cotyledon stages, and S17l aligns with the maturation stage. During S15, RECs exhibit mesophyll-like characteristics (cell state 1), lose their photosynthetic properties, and move to transition cell state 2. After completing floral organ abscission at S16 and the onset of longitudinal elongation of the silique at S17e, state 2 RECs differentiate into epidermal cells. Eventually, the floral abscission zones become covered by cuticle layers (cell state 3, S17m). RECs are depicted in pink, while SECs are in black. The flower development stages and their durations (h) are referenced according to Alvarez-Buylla et al.32. b, Heatmap showing the dynamic expression of transcription factors specific to each state over pseudotime. The color scale represents relative gene expression levels. The same cell state color code is used for all main and extended data figures.
Extended Data Fig. 6 Expression pattern of MYB74 and abscission-related genes in the AZ.
a-b, Spatiotemporal expression of prMYB74:nlsGFP-GUS in AZs: (a) the Z-projected surface scan view from S15 to S17l and (b) the transverse section view from S14 to S17m. In b, the top panel shows GUS expression in RECs, and the bottom panel shows GFP expression in the receptacle from which images were obtained using light-sheet microscopy. Scale bars, 50 µm (top) and 100 µm (bottom). c, Expression of abscission-related genes (BOP2, HSL2, and QRT2) in multiple cell layers of AZs before and after abscission. Scale bar, 50 µm. d, Expression of MYB74 in the transverse and longitudinal sections from the abscission-defective mutants hae hsl2 and ida, respectively. Scale bar, 50 µm.
Extended Data Fig. 7 RNA-seq analysis of MYB74-induced transcriptomic changes in AZ and receptacles shows that MYB74 controls the expression of genes associated with REC transdifferentiation.
a, RT-qPCR results show that MYB74 is induced following oestradiol treatment compared to mock treatment (Mock) and that the induction of MYB74 upregulates genes involved in the synthesis of cutin precursors. For all analyses, Actin2 was used as an internal control. The fold change was calculated relative to the mock-treated samples at 2 hours. The data shown were mean ± SD from three biological repeats. b, Heatmap displaying differentially expressed genes (DEGs) between oestradiol- and mock-treated samples (three biological replicates each). Statistical significance was evaluated using a two-sided Student’s t-test and graph showing all DEGs that met specific cut-off values (that is, an adjusted p < 0.05 and log2 fold change > 2). c, Heatmap showing the differential expression of genes related to photosynthesis and cuticle upon the induction of MYB74. d, Venn diagram showing that many REC-expressed MYB74 target genes (that is, 687 of 2176) show responses to MYB74 induction, with 272 and 415 being significantly up- and down-regulated, respectively. e, Gene ontology (GO) enrichment analysis for REC state-specific MYB74 target genes (2,176). Dot size represents the number of cells found to express a given gene, and its color represents its adjusted p-value (corrected for false discovery rate).
Extended Data Fig. 8 MYB transcription factors are expressed in RECs, rendering functional redundancy.
a, FA plots showing the expression of MYB family members enriched at the transition stage. The color scale represents the relative expression level. b-c, Surface-scanned fluorescent images of AZs (b) and longitudinally-sectioned images (c) reveal that MYB102 and MYB2 exhibit spatiotemporal expression similar to MYB74. Scale bar, 100 µm (b) and 50 µm (c). d, Loss-of-function mutations of myb74, myb74 myb102, and myb74 myb2 myb102 do not significantly affect cuticle layer formation. Cuticle formation and integrity were assessed by toluidine blue (TB) staining. Stage-17 siliques of the primary inflorescence at each flower position were analyzed. A representative result from three independent tests ( > 5 primary inflorescences for each test) is presented. Scale bars,100 µm. e, The first silique position that had a significant reduction in TB staining. TB-stained areas were quantified using Image J from acquired images in (d), and the position of the first silique where the stained area dropped to below 25% of the total AZ area was identified. The representative result is presented as the mean ± SD (n = 10). Statistical significance was determined using one-way ANOVA followed by Fisher’s test. The letters indicate no significant difference between genotypes (p = 0.8573).
Extended Data Fig. 9 MYB74 plays a role in REC transdifferentiation and cuticle layer formation.
a, Schematic representation of the prMYB74:MYB74-SRDX construct used to express the chimeric repressor MYB74-SRDX. The dark box indicates the SRDX fragment, fused to the MYB74 coding sequence (CDS). Blue arrows indicate the locations of primers used for RT-qPCR analysis of MYB74-SRDX expression. b, Compared to wild-type plants, the relative expression levels of MYB74-SRDX in prMYB74:MYB74-SRDX plants were examined using RT-qPCR assays. Total RNA was extracted from the receptacles of T2 plants from two independent transgenic lines (that is, #5 and #35). UBQ10 was used as an internal control. The error bars indicated mean ± SD from three biological replicates. c, Scanning electron microscope (SEM) images show the AZs and RECs’ surfaces of S17m siliques. In WT, an intact cuticle layer has formed, effectively filling the gaps between RECs. However, epidermis-specific expression of the cutinase gene CUTICLE DESTRUCTING FACTOR1 (CDEF1) impairs cuticle formation, resulting in unfilled gaps (indicated by an arrowhead). This plant line was able to flower but did not produce normal flowers. Scale bars, 50 µm (top) and 5 µm (bottom). d, Dual luciferase assay shows that MYB74 directly regulates GPAT4 promoter activity. A vector containing the multicloning site instead of the MYB74 coding sequence was a control. The error bars showed mean ± SD from three biological replicates. e, Co-expression of MYB74 and GPAT4 in the outermost layer of RECs, which differentiate into epidermal cells. Note the presence of inner RECs expressing only MYB74. Scale bars, 25 µm. f, Altered expression of the epidermal marker gene ATML1 in the RECs of plants carrying prUBQ10 > >MYB74-SRDX and prATML1:nls-GFP following MYB74-SRDX induction via one-day treatment with ß-estradiol. Scale bar, 50 µm.
Extended Data Fig. 10 The repressed function of MYB74 affects both receptacle and fruit growth.
a, Representative transverse section images of WT, MYB74-SRDX #35, and MYB74-SRDX #5 receptacles using resin embedding. These show the anatomical changes in transgenic plants, including reduced diameters, enlarged epidermal RECs, and cortical abnormalities. Scale bar, 100 µm. b, The distance from the pith center to the AZ vasculature was measured from section images in (a): Three consecutive 2 μm-thick transverse sections were taken from each receptacle sample. In each section, 1 to 4 sepal abscission zones were identified, each displaying lateral vasculatures. The radius was measured as the distance from the center of the pith to the sepal AZ vasculature (black lines). The mean radius from all measurements for each receptacle was then calculated. Five receptacles, each containing four sepal abscission zones, were examined for each genotype. From the 20 sepal abscission zones, 114 measurements were made from the section images for WT, #35, and #5 each. Box limits represent the 25th to 75th percentiles, whiskers show the entire data range, and the center line indicates the median. Statistical significance was determined via one-way ANOVA with Fisher’s test. c, Relative expression analysis showed that the induction of MYB74 by oestradiol upregulates five target MYB genes that negatively modulate lignification. Expression levels, based on TPM (Transcripts Per Million) were presented as mean ± SD from biological triplicates. d, Comparison of primary inflorescence stems between WT plants and transgenic plants carrying prMYB74:MYB74-SRDX. Arrowheads indicate siliques that are smaller than WT siliques. Scale bar, 1 cm.
Supplementary information
Supplementary Table 1
Supplementary Data S1: List of MYB74 target genes that are expressed in RECs and downregulated or upregulated in response to β-oestradiol induction.
Supplementary Table 2
Supplementary Data S2: List of putative target genes of MYB74 that are expressed in RECs.
Supplementary Table 3
Supplementary Data S3: List of primers used for this research.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wen, X., Lee, C.W., Kim, S. et al. MYB74 transcription factor guides de novo specification of epidermal cells in the abscission zone of Arabidopsis. Nat. Plants 11, 849–860 (2025). https://doi.org/10.1038/s41477-025-01976-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-01976-0
This article is cited by
-
Technovit Sectioning for High-Resolution Visualization of Tissue Anatomy and Gene Expression in the Floral Abscission Zone of Arabidopsis
Journal of Plant Biology (2026)
-
Making a new epidermis after abscission
Nature Plants (2025)
-
Heal the wound to make a better seal
Nature Plants (2025)
-
MYB Transcription Factors in Plant Developmental Plasticity
Journal of Plant Biology (2025)