Abstract
Complex molecules-mediated interactions, which are based on the bidirectional information exchange between microbes and human cells, enable the defense against diseases and health maintenance. Recently, diverse single-direction interactions based on active metabolites, immunity factors, and quorum sensing signals have largely been summarized separately. In this review, according to a simplified timeline, we proposed the framework of Molecules-mediated Bidirectional Interactions (MBI) between microbe and humans to decipher and understand their intricate interactions systematically. About the microbe-derived interactions, we summarized various molecules, such as short-chain fatty acids, bile acids, tryptophan catabolites, and quorum sensing molecules, and their corresponding human receptors. Concerning the human-derived interactions, we reviewed the effect of human molecules, including hormones, cytokines, and other circulatory metabolites on microbial characteristics and phenotypes. Finally, we discussed the challenges and trends for developing and deciphering molecule-mediated bidirectional interactions and their potential applications in the guard of human health.
Similar content being viewed by others
Introduction
The human microbiome consists of trillions of microorganisms, including a variety of viruses, bacteria, and fungi. They play a vital role in developing the immune system, maintaining cellular immune homeostasis, and guarding human health1,2,3,4,5. Microbes can influence the human physiology through active metabolic molecules, quorum sensing (QS), and other effectors6,7,8 and active metabolic molecules. Human cells in turn provide food and shelter for microbes and affect their growth and virulence through hormones, cytokine, and signaling metabolites9,10,11,12,13. Thus, microbe-human interactions include a high degree of crosstalk among different signaling pathways, such as QS systems14,15,16,17.
QS is the basis of the microbial sophisticated regulatory or signaling functions, such as competence, conjugation, biofilm development, and virulence18,19. Increasing evidence suggests that QS molecules are not only used for communications among microbes, but also for microbe-host interactions19,20,21. For example, Moura-Alves et al.22 showed that the concentration of acyl-homoserine lactones (AHLs), quinolones, and phenazines from Pseudomonas aeruginosa could be monitored by the aryl hydrocarbon receptor (AhR) of the host cells (zebrafish, mice, and human cells) to tune the corresponding immune responses dynamically. Recently, diffusible signal factors (DSFs) have also been reviewed to mediate the inter-kingdom communication among microbes, fungi (such as Candida albicans), plants, insects, and zebrafish22,23,24. Zhu et al.23 demonstrate that the anti-inflammatory effect of the DSF might be through Toll-like receptor signaling to attenuate the expression of inflammatory factors and lysosome-mediated apoptosis. The eavesdropping microbe-host communications by promiscuous receptors are very important for microbial interactions25,26. Typically, there is communication between autoinducer 3 (AI-3) and human hormones (epinephrine and norepinephrine)27 which can participate in the expression of microbial virulence factors and the regulation of the human immune system28. Furthermore, dynorphin and ethanolamine, two common metabolites in the mammalian gut, can also activate the QS-related quinolone signaling molecules of P. aeruginosa29 and Vibrio cholerae30, thus enhancing the microbial virulence and the colonization. Furthermore, Urbano et al.31 also noted that host-derived nitric oxide would disrupt microbial communication by targeting on the cysteine residue of AgrA in Staphylococcus aureus. Jugder et al.32 demonstrated that the QS regulator HapR of V. cholerae could stimulate the production of host-derived serotonin, thus activating gut immunity to promote host survival.
Certainly, to decipher the complex microbe-human interactions, some good reviews have already linked human cells with the microbiota by comprehensive consideration of active molecules33,34,35,36,37,38, immunity39,40,41,42, diseases14,43,44, and QS systems6,20,45,46. However, most research only summarized molecules derived from microbiota regulating the human phenotype in a single-direction, and few studies focused on human-derived molecules reversely shaping the microbiota. Even though some lectures have summarized the interactions based on human-derived molecules, most of them focus more on microbial effects on health and diseases, but the mechanisms and receptors involved are often overlooked.
To sum up, in this review, combining the microbial and human-derived molecules, we proposed the framework of the Molecules-mediated Bidirectional Interactions (MBI) between microbes and human cells, to systematically understand their intricate interactions. Note that there are lots of molecules involved in microbe-human interactions. It is impossible to give a full description of every molecule, we instead give a summary of the updated cases, and then propose a frame to enhance the understanding of MBI. Therefore, we mainly focused on molecules that participate in bidirectional interactions and possess known or potential inter-kingdom receptors. With regard to the interaction between microbes to human cells, we summarized different microbe-derived molecules and offered further insights into how the microbes affect the biological activities of different human cells. With respect to the interactions from human cells to microbes, we reviewed the effect of human-derived molecules on the characteristics and phenotypes of diverse microbes.
A timeline of microbe-human interactions
To better understand the interactions between microbes and human cells, we have provided a timeline to summarize the selective highlights for the development of microbe-human interactions since the 21st century (more details in Fig. 1)29,30,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82.
On the one hand, microbes communicated with human cells through various molecules14,58,83,84,85, such as short-chain fatty acids (SCFAs)33, tryptophan catabolites51,86 secondary bile acids (BAs)37,57 and QS molecules49,52. Most molecules affected human physiology by targeting different human receptors87, such as peroxisome proliferator-activated receptor (PPAR), G protein-coupled receptor (GPCR)83,88, AhR89, and T cell receptor (TCR) (more details in the left panel of the Fig. 1). On the other hand, some human-derived molecules participated in regulating microbial growth, virulence, or other physiologic functions90. These molecules contained hormones (such as dynorphin91 and catecholamines), cytokines (such as interferon γ, tumor necrosis factor α, and interleukin-1)69, and some circulatory metabolites (such as itaconate79 and succinate62). Among them, epinephrine, norepinephrine92, ethanolamine30, and serotonin93 could be recognized and bonded by different microbial QS receptors (more details in the right panel of Fig. 1)77.
The proposed timeline comprehensively summarized the development of the bidirectional interactions between microbes and human cells. As indicated in Fig. 1, the bidirectional promiscuous receptors of different molecules are receiving increasing attention and discovery, which will be beneficial for accurately and profoundly understanding the complex microbe-human interactions. In the following two parts, we will introduce the details of bidirectional interactions between microbes and human cells based on diverse molecules.
The interactions mediated by microbe-derived molecules
The interactions between microbes to human cells are mainly mediated by various microbial molecules. In this section, to give a broad illustration of different molecules and mechanisms, we would like to mainly summarize the interactions mediated by microbe-derived molecules, including short-chain fatty acids (SCFAs), bile acids (BAs), tryptophan catabolites, QS molecules, and others.
Short-chain fatty acids (SCFAs) -mediated interactions
Specifically, SCFAs are the most well-studied gut microbe-derived molecules with fewer than six carbons in the aliphatic tail94. The major SCFAs formed by the gut microbes are acetate (C2), propionate (C3), and butyrate (C4), which account for approximately 80% of all SCFAs, and the molar ratio of acetate, propionate, and butyrate in the colon is approximately 3:1:1, respectively34,35,95. SCFAs are known to affect human cells via the activation of several host receptors33. Currently, there are some well-characterized human SCFA-sensing G-protein-coupled receptors (GPCRs)96 which express on intestinal enterocytes and other cells of organs and tissues such as the liver, muscle, enteric neurons, and immune cells–indicating the breadth of their potential interactions throughout the human body. Among them, GPCR43 (also named as FFAR3) recognizes all three major SCFAs, but the affinities for GPCR41 (also named as FFAR2) are in the order of propionate, butyrate, and acetate, whereas GPCR109A (also named as HCAR2, NIACR1) interacts only with butyrate46,76. Apart from the three best-studied SCFA receptors, odorant receptors (ORs) belong to GPCRs, and OR51E2 residents in lung cells and also show ligand selectivity and sensitivity to the acetate and propionate97. Moreover, olfactory receptor 78 (Olfr78) is activated by acetate and propionate but not by butyrate, the EC50 values are 2.35 mM and 920 μM, respectively. The binding of gut microbiota-derived SCFAs to Olfr 78 induces the release of renin, which is involved in the modulation of blood pressure50. More details about the human receptors activated by SCFAs are listed in Table 1. The aforementioned findings suggest that SCFAs can affect the metabolism and immunity of human cells through different mechanisms with diverse receptors.
Bile acids (BAs) -mediated interactions
Bile acids (BAs) are amphipathic cholesterol metabolites37,57. The BA pool consists of primary BAs and secondary BAs. Primary BAs are synthesized from cholesterol in the liver, then the gut microbiota metabolize BAs secreted into the duodenum into secondary BAs. Besides the digestive functions, BAs exert diverse actions on the metabolism and immunity through human BAs receptors98. The Farnesoid X receptor (FXR)99 and the Takeda G protein-coupled receptor 5100 (TGR5; also known as G protein-coupled bile acid receptor 1, GPBAR1) are the two most studied bile acid receptors. Other receptors can also be directly or indirectly activated by bile acids, including the Shingosine-1-phosphate receptor 2 (S1PR2)99, the pregnane X receptor (PXR)101,102, the constitutive androstane receptor (CAR)103, the vitamin D receptor (VDR)104, and the retinoic acid-related orphan receptor γt (RORγt)105.
Tryptophan catabolites-mediated interactions
The microbial tryptophan catabolites play potential roles in mediating microbe-human interactions106. The microbial metabolites of tryptophan include indole and its derivatives, as well as tryptamine, all of which are aryl hydrocarbon receptor (AhR) ligands86. Indole is able to modulate the secretion of glucagon-like peptide-1 (GLP-1), which plays a critical role in stimulating insulin secretion107. Indole-based interactions can enhance gut epithelial barriers, elicit proinflammatory effects, and influence the immune response108. Specifically, Kumar et al.109 found that indoles from the commensal microbiota bind with AhR and tune the cellular composition of the colonic epithelium during aging. Indole-3-propionic acid (IPA) serve as an endogenous ligand for PXR, which strengthens the gut barrier through toll-like receptor 4 (TLR4) signaling or by inducing epithelial IL-10 receptor 1. Moreover, both of indole and its derivatives enhance IL-10 expression through AhR, which is linked with barrier function90. Tryptamine was found to accelerate transit time in vivo through the 5-HT4 receptor (5-HT4R), it is a GPCR uniquely expressed in the colonic epithelium34. Therefore, tryptophan catabolites may modulate the enteroendocrine system, metabolic homeostasis, and immunity of human cells106.
QS-mediated communication
QS, a vital microbial cell-to-cell communication system, is maintained by different signaling molecules that control phenotypic traits, virulence factors, biofilm formation, and dispersion19,110. QS was initially described in various microbes18, more emerging evidence indicated that pathogens applied QS systems to crosstalk human hormones, immunity factors56, and metabolites as cues to survival in the human environment55.
Indeed, some QS molecules were detected by humans and provoked specific immune responses. For example, AHLs, a classical type of QS molecules (QSMs), have been demonstrated to have immunomodulatory activities through boosting the production of IL-8 and inhibiting of lymphocyte proliferation, TNF, and IL-12111. Specifically, Jahoor et al.48 found that the transcriptional activity of PPARγ was inhibited by oxo-C12-HSL, while the activity of PPARβ/δ was increased. Moura-Alves et al.55 reported that several classes of P. aeruginosa QSMs, including HSLs (e.g., N-3-oxo-dodecanoyl-homoserine lactone), quinolones (e.g., 4-hydroxy-2-heptylquinoline), and phenazines (e.g., pyocyanin) can be detected by the AhR of zebrafish, mice, and human cells in a qualitative and quantified way. Pundir et al.56 found that human mast-cell-specific G-protein-coupled receptor MRGPRX2 and its mouse homolog Mrgprb2 are receptors for a part of Gram-positive QSMs (competence-stimulating peptide, CSP-1), triggering mast cell degranulation, which inhibited microbial growth and prevented biofilm formation. Legionella automatic inducer-1 (LAI-1, 3-hydroxypentadecane-4-one), a QS molecule of opportunistic pathogenic bacterium Legionella pneumophila, was employed for pathogen-human interaction52. For example, LAI-1 could modulate the migration of eukaryotic cells through a signaling pathway involving the scaffold protein IQGAP1, the small GTPase Cdc42 and the Cdc42-specific guanine nucleotide exchange factor ARHGEF9.
To sum up, diverse QS signals involve the mechanisms of crosstalk between the microbes and human cells91, such as the crosstalk among the microbial autoinducer-3 (AI-3) and human-derived epinephrine and norepinephrine21.
Other molecules-mediated interactions
Except for the aforementioned typical molecules, many other microbial molecules also have the capacity to communicate with human cells, such as the second messengers and some other active metabolites.
Some other active metabolites could affect the immune responses and characteristics changes of human cells. Specifically, Chen et al.112 identified that human gut microbes can secrete active small molecules (such as histamine, phenylethylamine, and amphetamine), which regulate human physiological and pathological activities by activating the corresponding GPCRs (such as histamine receptors, dopamine receptors, GPR56/AGRG1 and GPR97/AGRG3). Recently, Chen et al.60 developed a highly multiplexed screening technology (PRESTO-Salsa) to screen the microbial GPCR agonists. They found that microbial culture supernatants tended to activate human receptors coupled with epinephrine, dopamine, formylpeptide, histamine and succinic acid.
Microbial second messengers cyclic diadenylate monophosphate (c-di-AMP) and cyclic diguanylate monophosphate (c-di-GMP) are ubiquitously expressed in microbial species but absent in higher eukaryotes. The c-di-AMP and c-di-GMP can influence microbial cell survival, differentiation, colonization, biofilm formation, virulence, and microbe–host interactions113. Recent studies demonstrated that microbial c-di-GMP or c-di-AMP in macrophages could activate type I IFN production. A stimulator of interferon genes (STING) was shown to directly bind c-di-GMP, and to elicit strong interferon responses. Therefore, the second messenger-based interactions have provided remarkable examples of pathogen-host interactions in which a unique microbial molecule directly engages the innate immune system114.
The human fungal landscape is less populated in the body, but can greatly affect the immune system recognition and memory, combating pathogenic microorganisms, while being involved in important physiological processes, including metabolic functions. There are over 101 species belonging to 85 fungal genera in the oral cavity of healthy people, whereas there are 66-247 genera present in the gut. Aspergillus, Candida, Cladosporium, Cryptococcus, Galactomyces, Malassezia, Saccharomyces, and Trichosporon are in the lower gastrointestinal tract. Among them, Malassezia spp. are prevalent on the skin, while Candida, Aspergillus, and Cladosporium spp. are the gut harbors. Recognition of Candida albicans is mediated by several classes of pattern-recognition receptors localized on the surface of myeloid cells, including C-type lectin receptors and Toll like receptors. C-type lectin receptors recognize pathogen-associated molecular patterns (PAMPs) such as mannans and β-glucans on the fungal cell wall, resultingly in triggering an intracellular “proinflammatory storm” to curtail fungal invasion115. Reported in the signal peptide CPL1 of the human pathogen Cryptococcus neoformans interacts with Toll-like receptor4 to enhance macrophage polarization, trigger allergic inflammation, and promote fungal infection116.
The last but not least, viral infections and their role in disease provide global insight into their possible significance. The human virome is vast and complex, including approximately 1013 particles in humans with great heterogeneity. Gut viruses could be involved in numerous ways with the maturation of the human immune system, either indirectly or directly117. The latter has been observed especially mediated by118 many extracellular vesicles17 and cytosolic nucleic acid sensors detecting viral genomic materials. Note that the TLR9 has been singled out to be a key receptor for the recognition of viral DNA. The pattern recognition receptors (PRRs) have been shown to recognize viral components. Otherwise, the RIG-I-like receptors (RLRs), and NOD-like receptors (NLRs) also participate in the process. It is suggested that a direct interaction is widespread between the virus and the human immune system, independent of the host bacteria119.
The interactions mediated by human-derived molecules
Accumulating evidence showed that the highly mutualistic relationships between microbiota and human cells destined for the occurrence of bidirectional information exchange11,14. Although there are numerous studies on microbe-derived interactions, of which corresponding mechanisms and receptors have been recognized (Table 1), the specific signaling pathways and mechanisms for human-derived interactions are still limited and separate. Therefore, in this section, to have a better understanding of human-derived interactions, thus refining the MBI framework, we will give an introduction for more details of each interaction from human cells to microbes, which are mainly mediated by hormones, immunity factors, and circulatory metabolites, etc.
Hormone-mediated interactions
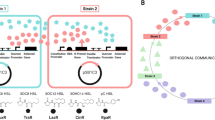
Much evidence proves the strong interconnections between hormones and microbiome91. Microbial endocrinology is a transdisciplinary discipline used to study the crosstalk mechanisms of microbes and human cells based on the hormones120,121. In fact, the human microbiota, a second endocrine organ122, have the ability to produce hormone-like molecules67. The prevalence of neuroendocrine hormones and their respective receptors in both human cells and microbes suggests the wide existence of the bidirectional interactions123 (more details in Fig. 2).
Hormones from various organs affect the different microbial phenotypes, such as the cell growth (representing by G), biofilms (representing by B) and virulence (representing by V). The colored triangle represents hormones that can bind to specific or unnamed potential microbial receptors. The color of triangle is consistent with the color of their corresponding microbial receptors. Grey receptors marked with “?” represent the unnamed potential microbial receptors that are speculated in the referenced literature. Hormones that are not labeled with colored triangles mean they affect the phenotype of microbes, but their potential microbial receptors have not been reported yet. Arrow indicates the promotion of microbial phenotypes, while the blunt arrow represents the inhibitions. The diagram was created by Figdraw (www.figdraw.com/).
Specifically, dynorphin91, a hormone belonging to the family of opioids, was reported to mediate interkingdom interactions. Dynorphin interfered with QS circuitry of P. aeruginosa and enhanced microbial virulence29. Dynorphin induced the expression of multiple virulence factor regulator (MvfR)–pqsABCDE and directly increased the production of three QS signals of P. aeruginosa, including 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), 4-hydroxy-2-heptylquinoline (HHQ), and Pseudomonas quinolone signal (3,4-dihydroxy-2-heptylquino-line, PQS). A reasonable hypothesis is dynorphin have an ability to bind intra-microbial opioid receptor-like molecules. However, until now, the cytoplasmic target of dynorphin in microbes has not been identified120. Additional opioid hormones, β-endorphin, and met-enkephalin, which were also involved in the stress response, also significantly increased the growth of E. coli in vitro124.
Natriuretic hormones possess a structure similar to cationic antimicrobial peptides and can be sensed by microbes73. This family contains three structurally related peptides, atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and C-type natriuretic peptide (CNP)92. Among them, BNP and CNP can increase the cytotoxicity and modify the lipopolysaccharide (LPS) of P. aeruginosa and P. fluorescens. Furthermore, both BNP and CNP significantly enhanced microbial production of the AHLs, as well as increasing the transcriptional level of AlgC that refers to its virulence73. Additionally, it was reported that serotonin activated the LasR QS pathway of P. aeruginosa at μM concentrations, then, it stimulated the production of microbial virulence factors and increased biofilm formation93. Serotonin could also be recognized and combined with QS regulator HapR of V. cholerae, which in turn activated host intestinal innate immunity to promote host survival.
Catecholamines contain epinephrine, norepinephrine, and dopamine, which are a type of typical human hormones72. Epinephrine and norepinephrine are the first human neurohormones affecting the QS system of microbe92. Dopamine, the immediate precursor of norepinephrine, is a kind of neurohormone within the gastrointestinal tract and stomach38,123. The identification of catecholamines receptor120 in enterohaemorrhagic Escherichia coli (EHEC) O157:H7 helps us further understand the human-microbe interaction72. QseC70, the receptor of autoinducer AI-3 in EHEC O157:H7, can also detect the human epinephrine, norepinephrine, and dopamine125. When these hormones bound with QseC, the phosphate moiety transferred from QseC to QseB, and then activated the transcription of the flagellar regulon. As a result, the microbial growth, motility, biofilm formation, adhesins and Shiga toxins in EHEC O157:H7 were increased38, which was a common feature of catecholamine hormones70. In addition, the opportunistic oral pathogen Aggregatibacter actinomycetemcomitans also employed QseC sensor kinase to interact with epinephrine from human neutrophils, resulting in the promotion of microbial growth and survival126. During the P. aeruginosa exposure to norepinephrine, the genes involved in the regulation of the virulence determinants pyocyanin, elastase, and the PQS were upregulated. Then, the virulence factor production, invasion of HCT-8 epithelial cells, and swimming motility were promoted in a concentration-dependent manner along with the increase of the transcriptional activation of LasR71. Furthermore, In Vibrio species, epinephrine, norepinephrine, and dopamine were found to promote the growth of Vibrio parahaemolyticus and Vibrio mimicu. The growth of Vibrio vulnificus was stimulated with high concentrations of epinephrine, indicating that the specificity and magnitude of the response of catecholamines varies among Vibrio species. As for Bifidobacterium angulatum GT102127, histidine kinase HK10 belongs to the family of kinases, which also serve as microbial epinephrine receptors72.
Gastrin, a peptide hormone secreted by stomach cells, stimulated the release of gastric acid and increased the growth rate of Helicobacter pylor92. Although the sensor for gastrin in H. pylori has not yet been identified, the fact that this response can be blocked by gastrin analogs suggests the presence of a microbial receptor for this hormone. It is also noteworthy that this response is restricted to human gastrin, and this may explain the specificity of H. pylori to humans. Another gastric peptide hormone, somatostatin, which inhibited the secretion of gastric acid, suppressed H. pylori growth. Somatostatin can directly bind to microbe, and this binding can be blocked by an anti somatostatin antibody, again suggesting that H. pylori harbored an as-yet-unidentified receptor for this peptide92. Moreover, H. pylori infection also increased and decreased the levels of gastrin and somatostatin, respectively. Insulin can bind and increase the growth and virulence of Burkholderia pseudomallei and the related species Burkholderia cepacia by reducing the activity of phospholipase C and acid phosphatase. However, the microbial insulin receptor remains unknown92.
Gut microbiota is also involved in maintaining the balance of estrogen metabolism. Note that the estrogen can indirectly regulate the gut microbiota composition and diversity128. More and more evidence has also found the effects of estradiol on the regulation of QS-related phenotypes129. For example, estradiol increased the attached biofilm mass and remodeled the biofilm ultrastructure of P. aeruginosa through the enhancement of coverage with extracellular polymeric substance and average surface roughness, respectively. QS activity measurements performed in biofilm supernatants via AHL reporter further showed that estradiol treatment modulates microbial QS systems indeed.
To sum up, human hormones, including neurohormones, digestive hormones, catecholamines, and estrogen, will indeed affect microbial characteristics and phenotypes, which provides a compelling explanation for their important role in the guard of human health. Most of the human hormones mentioned above have positive effects on the microbial growth or virulence, which indicates that the human body may be more susceptible to infection with pathogens in cases of endocrine disorders. Although some QS receptors have been recognized, the corresponding receptors of the regulatory mechanisms remain vague (represented by the question mark in the Fig. 2), which needs more attention and future research.
Cytokines-mediated interactions
The immunity system is the main battlefield for negotiations between human cells and microbes based on cytokines38,130,131,132, which are effective mediums for humans in response to microbial attacks74. Although human cells are known to express different receptors that bind microbes for the purpose of activating the immune system, it must be considered that microbes themselves might possess specialized receptors that in turn recognize and respond to human immune (more details in Fig. 3)69. For example, B. angulatum GT102 not only possessed microbial epinephrine receptor as mentioned above, but also equipped a fibronectin type III domain-containing protein having cytokine receptor motifs, which was possibly capable of interacting with human cytokines127. A number of studies have also reported the ability of cytokines to bind to microbes, and this was frequently associated with altering microbial phenotypes, such as cell growth and biofilm. In this section, we will provide a summary of human-derived interactions based on some typical cytokines, such as different interleukins, tumor necrosis factor (TNFs), and interferon-γ (IFN-γ).
The colored triangle represents human cytokine that can bind to specific or unnamed potential microbial receptors. The labeling principles for Microbial receptors are the same as those in Fig. 2. The microbe involved in this figure exist in the human mouth, lungs, stomach, and gut. Human cytokines from immunity cells exert antimicrobial effects by targeting microbial receptors. However, not all of binding can effectively combat pathogens, the virulence and growth of the pathogen may be activated by human cytokines. Arrow indicates the promotion of microbial phenotypes, such as the growth and virulence, while the blunt arrow represents the inhibitions. The diagram was created by Figdraw (www.figdraw.com/).
Specifically, interleukin-I (IL-1) is a polypeptide cytokine that mediates different infection and inflammation. The positive effect of IL-1β on microbial growth occurred in S. aureus. In addition, the opportunistic oral pathogen A. actinomycetemcomitans employs several specific strategies for enhancing its survival in the host: it can secrete a leukotoxin that directly targets human phagocyte cells, monocytes, and macrophages, and the cytolethal-distending toxin indirectly modulates periodontal bone resorption as well as periodontal keratinocyte and fibroblast proliferation75. It is possibly promoted the formation of biofilm and adjusted the virulence properties after sensing inflammation-related environmental factors through a specific receptor binding with IL-1β. Thus, the discovery of the microbial outer membrane IL-1β receptor was crucial for explaining the role of sensory cascade in the virulence of this opportunistic pathogen75. Similarly, Porat et al.133 found human IL-1β bound with virulent Escherichia coli in a specific and saturable fashion and then enhanced the growth of virulent E. coli, which was inhibited by IL-1 receptor antagonist. Thus, IL-1β may recognize a functional IL-1-like receptor structure in virulent E. coli and serve as a virulence factor for microbial pathogenicity134. The IL-18 triggered the growth of gut microbiota, it could bind with the lipopolysaccharide (LPS) of A. actinomycetemcomitans135. IL-8 increased E. coli adhesion and transmigration in the lung by binding with the Tsr receptor, a major methyl-accepting protein136.
TNFs are another typical cytokine, which primarily maintains the survival and balance of immune cells and usually includes tumor necrosis factor-alpha (TNF-α) and tumor necrosis factor beta (TNF-β). Tumor necrosis factor-α (TNF-α) plays an important role in innate immunity. Some in vitro studies have shown that TNF-α may serve as a growth factor for some microbes. The human-restricted pathogen Neisseria meningitidis, a major cause of microbial meningitis and sepsis worldwide, could bind with TNF-α or IL-8 through pilus assembly proteins (i.e. PilQ and PilE proteins), thus increasing its virulence137. Jin-Hwa Lee et al.138 found that recombinant mouse TNF-α increased in vitro proliferation of E. coli in a concentration-dependent manner, and this effect was attenuated by anti–TNF-α antibodies. The same E. coli growth-promoting effect was not observed with P. aeruginosa, which raised the possibility that E. coli may express an as yet unidentified receptor for TNF-α. Furthermore, Luo et al.139 have also reported that TNF-α can bind to E. coli, S. typhimurium, and Shigella flexneri. Particularly, they found that S. flexneri had 276 receptors for TNF-α with a Kd of 2.5 nM, and the invasion ability was enhanced after binding with them. Just like other gram-negative pathogen, S. flexneri has some receptors for TNF-α.
Interferon-γ (IFN-γ), another typical cytokine, could also contribute to the control and regulation of human-derived interactions. For instance, Wu et al.69 demonstrated a link between the QS system and interferon-γ (IFN-γ) in P. aeruginosa. Specifically, IFN-γ bound with an outer membrane protein OprF, resulting in the rhlI gene expression and increased production of C4-homoserine lactone (C4-HSL) as well as a QS dependent virulence determinant, the type I P. aeruginosa lectin140. When IFN-γ activated the QS system, it also enhanced the production of virulence factor pyocyanin, which most likely was required for the full virulence of P. aeruginosa in lung infections. Another pathogen Mycobacterium tuberculosis also had the ability to sense IFN-γ via the MmpL10 protein, leading to transcriptional changes that may enhance its virulence and accelerate growth81. The prediction result of the Alphafold model suggested a potential binding pose in the membrane-proximal area of the second MmpL10 extracellular region, which was consistent with the fact that IFN-γ binds the microbial cell surface. Furthermore, H. pylori -infected gastric mucosa was characterized by high levels of IFN-γ, but whether IFN-γ regulated the virulence of H. pylori was unclear. Afterward, Zhao et al.141 characterized the binding response of H. pylori to IFN-γ, which altered the expression of 14 proteins, including the virulence factor. The transcription and translation of cytotoxin-associated gene A (CagA) were downregulated coupled with the decreased degree of cell damage when co-cultured the human cell line AGS with H. pylori exposed to IFN-γ. Thus, IFN-γ contributed to control H. pylori infection indirectly through the effect on virulence factor CagA.
Note that only IFN-γ helps to inhibit the pathogen H.pylori, on the contrary, many other cytokines mentioned above encourage the growth or virulence of pathogens. This unexpected phenomenon reminds us that the protection of human cytokines from being stolen and utilized by pathogens is necessary when developing antimicrobial drugs. Many cytokines were reported to regulate the composition of gut microbiota, such as IFN-β, IL-33, IL-21, and TGF-β, while the corresponding microbial receptor information had not been reported39. Certainly, more specific mechanisms between the cytokines and microbial receptors are needed to identify to reveal the important role of cytokine sensing in the control of microbial physiological activities, such as biofilm142 and virulence140. In the future, we should focus on the identification of compounds structurally similar to cytokines that could modulate the expression of microbial virulence genes and reprogram the microbial behavior towards less pathogenic and more commensal states. This knowledge could be directed towards the study of drugs and existing inhibitors to assess their suitability as novel therapeutic agents. By exploiting knowledge of their modes and sites of action to combat microbial infections, it is possible to develop powerful novel arsenals against the serious problem of antibiotic resistance137.
Active metabolites-mediated interactions
In theory, cells are equipped with machinery for metabolite sensing, which enables them to synchronize metabolic processes with cell signaling and gene expression. Thus, a part of human-derived metabolites are important not only as nutrient intermediate but also as signaling molecules11. In this section, we will focus on the summary of signaling metabolites-mediated interactions based on the components in the tricarboxylic acid cycle (TCA) and other active bioactive molecules, such as ethanolamine, D-serine, and gamma-aminobutyric acid (GABA) (Fig. 4).
The colored hemisphere represents small molecule active metabolites in the human body. The labeling principles for microbial receptors follow Fig. 2. The red line represents the metabolic process centered around the TCA cycle, which converts the three major components of food (glucose, lipid, and protein) into small molecules. A part of molecules plays a signal role by binding to known/unknown microbial receptors. Arrow indicates the promotion of microbial growth or virulence. The diagram was created by Figdraw (www.figdraw.com/).
Specifically, some researchers have reported that the human-derived lactate played an essential role in the interactions between human cells and microbes, such as N. meningitidis143, The human pathogen N. meningitidis is the leading cause of pyogenic meningitis. The pathogenicity of N. meningitidis is related to the spread of microbial colonies. It was demonstrated that direct contact with host cells is not required for microcolony dispersal, instead of accumulation of a host-derived lactate. Interestingly, this mechanism occurs independently of metabolic utilization of the lactate by the microbes, suggesting that lactate possibly plays a role as a signaling molecule although the microbial receptor of lactate has not been found. Similarly, Neisseria gonorrhoeae microcolony dispersal could also be induced by lactate. These findings reveal the role of host-secreted lactate in microcolony dispersal and virulence of pathogenic Neisseria143. These findings will assist in better understanding the transition from asymptomatic carriage to invasive disease.
Itaconate is one of the most abundant metabolites produced by myeloid cells in response to infection and a major activator of the host antioxidant response. Recently, it has gradually been regarded as one of the important human-derived metabolites for the host-microbe interactions144. For example, Wang et al.78 discovered that the M. tuberculosis resisted the itaconate from macrophages by an essential bifunctional enzyme Rv2498c. Sebastián A. Riquelme et al. found that the itaconate-dominated immune metabolic response can lead the pathogen P. aeruginosa79 and S. aureus145 to change their metabolism to downregulate LPS and upregulate extracellular polysaccharides (EPS), thus promoting biofilm formation145. More specifically, P. aeruginosa induces the production of host-derived itaconate. The release of itaconate subjects P. aeruginosa to a major selective pressure. Meanwhile, itaconate selects metabolically adapted P. aeruginosa strains through the alteration of microbial metabolic activity and the increase of extracellular polysaccharides (EPS), thus increasing microbial resistance to oxidant stress. EPS not only protects the microbes from oxidants, but triggers myeloid cell metabolic reprogramming, both locally and in circulating monocytes, to induce even greater itaconate release79. Following a similar approach, using protein extracts, Sebastián A. Riquelme et al.145 further confirmed itaconate inhibited the aldolase activity of S. aureus in a dose-dependent manner. This glycolytic inhibition limited S. aureus planktonic growth, which indicated that itaconate induces metabolic stress in S. aureus by targeting glycolysis.
C4-dicarboxylates (aspartate, fumarate, malate, and succinate) are derived from the carbohydrates, proteins, and fats through the TCA cycle or some steps metabolic strategy of the reverse TCA cycle62. C4-dicarboxylates both serve as a nutrient source and as active metabolites for enteric microbes. Thus, they play a major and specific role in the interaction of the mammalian host with the pathogen. E. coli and S. typhimurium which are mainly use the two-component system DcuS/DcuR and the chemotaxis receptor Tar to perceive the C4-dicarboxylates. Specifically, L-Aspartate was important for the gut colonization of E. coli and S. typhimurium due to its role in fumarate respiration, nitrogen assimilation, and regulation of virulence146,147,148. Host- and microbiota-derived L-malate was sensed by DcuS/DcuR of EHEC O157:H7 during infection. Then, the L-malate was transduced to the master virulence regulator Ler, which activated the locus of enterocyte effacement (LEE) genes to promote adherence82. Succinate regulated the polarization of macrophages toward the M1 macrophages, which secreted proinflammatory cytokines (e.g., IL-6, IL-1β, and IFN-γ) as antimicrobial effectors to inhibit the pathogen’s spread. Note that Rosenberg et al.80 found that succinate accumulated in macrophages during the S. typhimurium infection. Intracellular S. typhimurium utilized human-derived succinate through the transporter DcuB and induced the expression of PmrAB and SPI-2 microbial virulence regulons to activate their survival mechanism. Thus, the power to accelerate pathogen growth in the human body may originate from the metabolic cycle system.
Certainly, in addition to the aforementioned active metabolites in the TCA cycle, some other bioactive human-derived molecules, such as ethanolamine30, D-serine149, GABA64,150, were also typical examples of active metabolites. Specifically, ethanolamine, a common intestinal metabolite, serves as a chemical source of QS interference30. When a V. cholerae mutant expressing only the histidine kinase QS receptor (CqsR) invaded the host, the inhibitory effect of ethanolamine on CqsR could be a possible source of QS interference but was masked by the presence of the other parallel QS pathways, allowing V. cholerae to robustly colonize the host30. For the D-serine, the EHEC can sense and transport the human-derived D-serine via the transporter its CycA protein, resulting in the inhibition of the microbial growth and type 3 secretion system-dependent colonization. Therefore, the corresponding disruption of transporters and/or activation of a previously nonfunctional D-serine deaminase will contribute to abrogating the inhibition of microbial growth149. As for GABA, microbes possess different sensor proteins that recognize specifically it—such as chemoreceptors151,152 and transporter153. Specifically, the GABA chemoreceptors in P. putida and P. aeruginosa bind their ligands with high affinity with respective KD values of 175 nM151 and 1.2 μM. Matilde Fernández et al.150 also found a GABA receptor PA0222 in P. aeruginosa, it was demonstrated that microbial chemotaxis to GABA ligand is due to the direct binding to PA0222. Furthermore, a three‐dimensional structure of specific GABA binding protein Atu4243, which affects the GABA transport and GABA-regulated functions, was identified in the Agrobacterium fabrum153. Taken together, aforementioned data indicate that microbes have evolved specific GABA binding proteins, belonging to different protein families that are involved in different interactions for phenotype control, such as chemotaxis.
To sum up, as summarized in Fig. 4, the components in the tricarboxylic acid cycle (TCA) (L-malate82, succinate, and itaconate79), ethanolamine30, and gamma-aminobutyric acid (GABA)64 were found to regulate the microbial characteristics by attaching to specific microbial receptors. Predictably, more and more active metabolites will be mined and proposed to mediate the microbe-human bidirectional interactions in the future.
Concluding remarks and future perspectives
Microbes and human cells have co-evolved for billions of years, through which they have been exposed to many molecules produced by each other. As summarized above, these molecules are important mediators for extra-and intracellular molecules. As illustrated in Fig. 5, microbes initiate interactions with human through diverse molecules, and their corresponding human receptors. More recently, these mechanisms have been described in the reverse direction. Humans employ hormones, cytokines, and other circulatory metabolites in response to microbes, which modulate the microbial characteristics and phenotypes. While detailed mechanistic studies linking specific microbial molecules to human physiological status has been deeply explored, the regulation of human molecules on microbial physiology and homeostasis is still nascent. In view of the fact that there are many advanced technologies, such as multi-omics and high-throughput screening approaches, various signaling pathways, and mechanisms will be deciphered and understood, thus the future molecules-mediated bidirectional interactions (MBI) will gradually enter the rapid development stage. Certainly, there will be more opportunities and challenges for further developing the MBI in potential applications of disease diagnosis and treatment.
Human cells seem to have inherited certain physiological mechanisms from microbes, such as the QS mechanisms. More and more researchers reasonably believe that QS is not restricted to microbiome55, some immune cells is working based on a mechanism similar to the QS system. For example, the IgM-secreting B cells ‘counted’ their own numbers, and IgG acts as the inducer that reports the density of B cells154. In the presence of antigenic stimulation, there was a concentration threshold for effector CD4+ T cells to secrete sufficient amounts of IL-2 to maintain STAT5 phosphorylation and the population expansion of effector T cells111. Similarly, the dendritic cell activation followed an all-or-nothing process based on a threshold level of type I IFN in the microenvironment155. Similar to type I IFN, the IL-10 was also found to be the QS inducer for dendritic cells. An increase in IL-10 concentration signaled to other dendritic cells to collectively decrease their pro-inflammatory cytokine gene expression111. In the immunity system, the cellular response will be predicted by the quorum of cytokine-producing cells and operate globally at the tissue level. These response modes are reminiscent of QS mechanisms where the accumulation of the autoinducer produced by many cells drives a collective response155. In summary, the human immune system may retain the QS mechanism of microbes. On the one hand, the activation mechanism of immune cells and secretion of cytokines are very similar to the microbial QS mechanism. On the other hand, cytokines can control pathogenic microbes by interfering with their QS system39. However, the receptors and their signaling pathways that the human cells use to profit themselves or eavesdrop on microbes remain poorly understood. More precisely, a question we are curious about is whether the human immune system evolved from the QS system of prokaryotes? The answer of this question will need more research about the definition and characteristics of QS systems.
The second clue is the chemical mimicry of the signal language between microbes and human cells. As we all know, AI-2 is a classic QS signal language in the microbial community. It was amazing that mammalian epithelial cells could release an AI-2 mimic, which was detected by the AI-2 receptor of enteric pathogen S. typhimurium and then stimulated the expression of QS-related genes77. Expect the human to mimic the microbial signal molecule, the microbe also learns the human biochemical molecules. Cohen et al.53 found that GPR119 agonists analog from commensal microbes regulate metabolic hormones and glucose homeostasis as efficiently as human ligands. AHLs, another type of representative QS molecules, are equipped with highly conserved serine-acylated. Coincidentally, the activity of human ghrelin is also related to serine-3 (Ser3) modified by N-octanoylated, which means the ghrelin was consistent with the characteristic of microbial AHL156. Sphingolipids, a lipid class characterized by a long-chain amino alcohol backbone, were first discovered in the human brain. They are structural membrane components and important eukaryotic signaling molecules. In recent research, more and more symbiotic microbes were found to produce sphingolipids, and the structure of microbial sphingolipids with those produced by humans is conservative157,158. Therefore, it is reasonable to presume that microbes and the human mimic each other’s interaction language, resulting in the improvement of structural homology between their signal molecules.
The MBI plays an important role in health maintenance and disease development. Microbes mainly reside in the human gut system and widely affect the whole body. Note that most of the MBIs collected in this review were the interactions between various bacteria and human cells. Although there was some evidence validated that small molecules from fungi and viruses (such as phages) would have also different interactions with the human immune cells (more details in Section “Other molecules-mediated interactions” and Table 1), the investigations for virus-derived or fungi-derived interactions with human cells are very limited. Given the widespread presence of fungi and viruses in the human gut microbiota, more researches are needed to focus on the elucidation of virus-human interactions and fungi-human interactions based on different interaction mechanisms in the near future. Only by incorporating phages, viruses, fungi, and bacteria into the gut microbial systems and studying their interactions with human cells can we understand the broader microbe-human interactions from a more complete perspective, as well as the prevention, control, and treatment of human diseases.
Furthermore, to understand the broader microbe-human interactions, the microbial extra-intestinal function shouldn’t be ignored. The interaction with organs beyond the gastrointestinal tract occurs directly via Toll-like receptors (TLR) and indirectly via different microbial signaling molecules159, some typical examples are as follows. Preclinical evidence has firmly established bidirectional interactions among the brain and the gut microbiome. Interaction within this system is nonlinear, is bidirectional with feedback loops, and candidate signaling molecules have been identified160. Gut microbiota affects the brain via the vagus nerve, cytokines, and their metabolites such as tryptophan, GABA, and acetylcholine. Meanwhile, many commensal and pathogenic microbes in the gut are sensitive to norepinephrine and dopamine, which were released from sympathetic postganglionic neurons into the gut lumen that can cause the expression of virulence genes, and increased growth of pathogens. With regard to the gut-lung axis, microbial SCFAs were reported to protect against allergic asthma, an effect that involved GPR41 signaling and reduced TH2 effector cell activation in the lung161. Moreover, maternal acetate, generated through microbial fermentation of dietary fiber can regulate gene expression in the lung through inhibition of histone deacetylase 9 (HDAC9) to protect the offspring against allergic asthma159. With respect to the gut-liver axis, the dysfunction of the gut mucosal barrier (“leaky gut”) and increased microbial translocation into the liver via the gut–liver axis probably play crucial roles in liver disease development and progression162. The gut liver axis is mainly the crosstalk between bile acids and gut microbiota, importantly it is a bidirectional interaction as the microbiota affects bile acid metabolism and bile acids affect microbiota composition.
The MBI framework we proposed is meaningful for understanding the complex microbe-human interactions. Certainly, there are some challenges in further understanding the MBI, mainly for the quantification and refinement of the reported systems as well as the discovery and analysis of the new systems. Note that metabolites can act as controllers of the phenotype — as opposed to being simply biomarkers of phenotypic states. Therefore, some metabolites serve as both energy conversion intermediates and inter-kingdom molecules, refreshing our traditional understanding of metabolites163. For example, aforementioned intermediates in the TCA cycle including L-malate, succinate, and itaconate, have been regarded as the signaling molecules and they regulate different microbial physiological activities. In addition, regulatory effects of some amino acids are gradually concern, such as serine. Therefore, we believe it is necessary to re-examine various small molecule metabolites from the perspective of signal interactions and explore their corresponding receptors. Active molecules serve as umbilical cords between microbes and the human body. Some of them most likely have come from a common ancestor or were appropriated by first eukaryotes from their pro-mitochondrial or pro-chloroplast symbionts. In other cases, horizontal gene transfer seems to be a plausible explanation. Anyway, one should not be shocked to find in a free-living bacterium a signaling domain seen previously only in eukaryotes. Conversely, many eukaryotic signaling domains appear to have roots in microbes. This is an unexpected but promising development, as data on eukaryotic signal transduction could help in deciphering the functions of microbial proteins and vice versa133.
The complex interactions between microbes and human cells require interdisciplinary research to unravel. In terms of methodology, the activity metabolomics can be conducted to identify the bioactive metabolites, which has soared above a purely biomarker-driven approach164. However, the activity screening guided by metabolomics needs to increase robustness, which can be achieved by combining metabolomics, systems biology, and bioactivity data. Activity metabolomics enables us to identify metabolites with the highest potential to modulate biological processes and cell physiology. Multi-omics techniques, as well as data mining and analysis based on artificial intelligence, will provide more clues for the mining of the potential interactions between microbes and human cells. In addition, network-based analysis approaches165 have the potential to promote the understanding of the function of the MBI in the identification of the relationships between active molecules crucial for community stability, and the prediction of the effects of various interactions on human health. To sum up, the manipulation of microbiota genes encoding metabolites that elicit host cellular responses will be a new small molecule therapeutic modality (microbiome-biosynthetic-gene-therapy)53. The prevalence of this ability suggests that microbe-human bidirectional interaction may be extremely common and that many more systems will be uncovered in the near future, which will contribute to more applications of microbiome research in personalized medicine and public health.
Data availability
No datasets were generated or analyzed during the current study.
Code availability
Code sharing does not apply to this article as no code was generated or used during the current study.
References
Aggarwal, N. et al. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 123, 31–72 (2022).
Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71 (2021).
Janney, A., Powrie, F. & Mann, E. H. Host-microbiota maladaptation in colorectal cancer. Nature 585, 509–517 (2020).
Cheng, A. G. et al. Design, construction, and in vivo augmentation of a complex gut microbiome. Cell 185, 3617–3636.e3619 (2022).
Ansaldo, E. & Belkaid, Y. How microbiota improve immunotherapy. Science 373, 966–967 (2021).
Wu, S., Xu, C., Liu, J., Liu, C. & Qiao, J. Vertical and horizontal quorum-sensing-based multicellular communications. Trends Microbiol. 29, 1130–1142 (2021).
Su, Y. et al. Bacterial quorum sensing orchestrates longitudinal interactions to shape microbiota assembly. Microbiome 11, 241 (2023).
Di Menna, L. et al. The bacterial quorum sensing molecule, 2-heptyl-3-hydroxy-4-quinolone (PQS), inhibits signal transduction mechanisms in brain tissue and is behaviorally active in mice. Pharmacol. Res. 170, 105691 (2021).
Xing, C. et al. Microbiota regulate innate immune signaling and protective immunity against cancer. Cell Host Microbe 29, 959–974 e957 (2021).
Behyar, M. B., Mirzaie, A., Hasanzadeh, M. & Shadjou, N. Advancements in biosensing of hormones: Recent progress and future trends. TrAC Trends Anal. Chem. 173, 117600 (2024).
Zhang, Y., Chen, R., Zhang, D., Qi, S. & Liu, Y. Metabolite interactions between host and microbiota during health and disease: Which feeds the other? Biomed. Pharmacother. 160, 114295 (2023).
Skwara, A. et al. Statistically learning the functional landscape of microbial communities. Nat. Ecol. Evol. 7, 1823–1833 (2023).
Zhernakova, D. V. et al. Host genetic regulation of human gut microbial structural variation. Nature 625, 813–821 (2024).
Lu, Q. & Stappenbeck, T. S. Local barriers configure systemic communications between the host and microbiota. Science 376, 950–955 (2022).
Wu, S. et al. Quorum sensing-based interactions among drugs, microbes, and diseases. Sci. China Life Sci. 66, 137–151 (2023).
Sam, S. A., Teel, J., Tegge, A. N., Bharadwaj, A. & Murali, T. M. XTalkDB: a database of signaling pathway crosstalk. Nucleic Acids Res. 45, D432–D439 (2017).
Huang, J. et al. Extracellular vesicles as a novel mediator of interkingdom communication. Cytokine Growth Factor Rev. 73, 173–184 (2023).
Wu, S. et al. Machine learning aided construction of the quorum sensing communication network for human gut microbiota. Nat. Commun. 13, 3079 (2022).
Wu, S., Liu, J., Liu, C., Yang, A. & Qiao, J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343 (2019).
Su, Y. & Ding, T. Targeting microbial quorum sensing: the next frontier to hinder bacterial driven gastrointestinal infections. Gut Microbes 15, 2252780 (2023).
Li, Q., Ren, Y. & Fu, X. Inter-kingdom signaling between gut microbiota and their host. Cell. Mol. Life Sci. CMLS 76, 2383–2389 (2019).
He, Y. W. et al. DSF-family quorum sensing signal-mediated intraspecies, interspecies, and inter-kingdom communication. Trends Microbiol. 31, 36–50 (2022).
Zhu, H. et al. Bacterial quorum-sensing signal DSF inhibits LPS-induced inflammations by Suppressing Toll-like receptor signaling and Preventing Lysosome-mediated apoptosis in zebrafish. Int. J. Mol. Sci. 23, 7110 (2022).
Yi, R. et al. Quorum-sensing Signal DSF inhibits the proliferation of intestinal pathogenic bacteria and alleviates inflammatory response to suppress DSS-induced colitis in Zebrafish. Nutrients 16, 1562 (2024).
Wu, S. et al. Design and analysis of quorum sensing language “Interpreter” ecosystem for microbial community. Chem. Eng. J. 496, 153148 (2024).
Wu, S., Qiao, J., Yang, A. & Liu, C. Potential of orthogonal and cross-talk quorum sensing for dynamic regulation in cocultivation. Chem. Eng. J. 445, 136720 (2022).
Karavolos, M. H., Winzer, K., Williams, P. & Khan, C. M. Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465 (2013).
Fan, Q. et al. Contribution of quorum sensing to virulence and antibiotic resistance in zoonotic bacteria. Biotechnol. Adv. 59, 107965 (2022).
Zaborina, O. et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 3, e35 (2007).
Watve, S. et al. Parallel quorum-sensing system in Vibrio cholerae prevents signal interference inside the host. PLoS Pathog. 16, e1008313 (2020).
Urbano, R. et al. Host Nitric Oxide Disrupts Microbial Cell-to-Cell Communication to Inhibit Staphylococcal Virulence. Cell Host Microbe 23, 594–606.e597 (2018).
Jugder, B. E. et al. Vibrio cholerae high cell density quorum sensing activates the host intestinal innate immune response. Cell Rep. 40, 111368 (2022).
van der Hee, B. & Wells, J. M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 29, 700–712 (2021).
Roager, H. M., Stanton, C. & Hall, L. J. Microbial metabolites as modulators of the infant gut microbiome and host-microbial interactions in early life. Gut Microbes 15, 2192151 (2023).
Dalile, B., Van Oudenhove, L., Vervliet, B. & Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478 (2019).
Zheng, X., Cai, X. & Hao, H. Emerging targetome and signalome landscape of gut microbial metabolites. Cell Metab. 34, 35–58 (2022).
Collins, S. L., Stine, J. G., Bisanz, J. E., Okafor, C. D. & Patterson, A. D. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat. Rev. Microbiol., 21, 236–247 (2023).
Zhang, C. et al. An overview of host-derived molecules that interact with gut microbiota. iMeta, 2, e88 (2023).
Zheng, D., Liwinski, T. & Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506 (2020).
Goguyer-Deschaumes, R., Waeckel, L., Killian, M., Rochereau, N. & Paul, S. Metabolites and secretory immunoglobulins: messengers and effectors of the host-microbiota intestinal equilibrium. Trends Immunol. 43, 63–77 (2022).
Garrett, W. S. Immune recognition of microbial metabolites. Nat. Rev. Immunol. 20, 91–92 (2020).
Michaudel, C. & Sokol, H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 32, 514–523 (2020).
Wu, J., Wang, K., Wang, X., Pang, Y. & Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 12, 360–373 (2021).
Lavelle, A. & Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237 (2020).
Zhang, Y., Ma, N., Tan, P. & Ma, X. Quorum sensing mediates gut bacterial communication and host-microbiota interaction. Crit. Rev. Food Sci. Nutr., 64, 3751–3763 (2024).
Xiao, Y. et al. Impact of quorum sensing signaling molecules in gram-negative bacteria on host cells: current understanding and future perspectives. Gut Microbes 14, 2039048 (2022).
Le Poul, E. et al. Functional Characterization of Human Receptors for Short Chain Fatty Acids and Their Role in Polymorphonuclear Cell Activation. J. Biol. Chem. 278, 25481–25489 (2003).
Jahoor, A. et al. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. Am. Soc. Microbiol. 190, 4408–4415 (2008).
Kravchenko, V. V. et al. Modulation of gene expression via disruption of NF-κB signaling by a bacterial small molecule. Science 321, 259–263 (2008).
Pluznick, J. L. et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl Acad. Sci. 110, 4410–4415 (2013).
Zelante, T. et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 39, 372–385 (2013).
Simon, S. et al. Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1-Cdc42-ARHGEF9-dependent pathway. PLoS Pathog. 11, e1005307 (2015).
Cohen, L. J. et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 (2017).
Elmanfi, S., Ma, X., Sintim, H. O., Könönen, E., Syrjänen, S. & Gursoy, U. K. Quorum-sensing molecule dihydroxy-2, 3-pentanedione and its analogs as regulators of epithelial integrity. J. Periodontal Res. 53, 414–421 (2018).
Moura-Alves, P. et al. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 366, eaaw1629 (2019).
Pundir, P. et al. A connective tissue mast-cell-specific receptor detects bacterial quorum-sensing molecules and mediates antibacterial immunity. Cell Host Microbe 26, 114–122.e118 (2019).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Schluter, J. et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 588, 303–307 (2020).
Hill, J. H. & Round, J. L. SnapShot: Microbiota effects on host physiology. Cell 184, 2796 (2021).
Chen, H. et al. Highly multiplexed bioactivity screening reveals human and microbiota metabolome-GPCRome interactions. Cell, 186, 3095–3110.e19 (2023).
Sanmarco, L. M., Rone, J. M., Polonio, C. M., Fernandez Lahore, L., Giovannoni, F. et al. Lactate limits CNS autoimmunity by stabilizing HIF-1α in dendritic cells. Nature. 620, 881–889 (2023).
Wei, Y. H., Ma, X., Zhao, J. C., Wang, X. Q. & Gao, C. Q. Succinate metabolism and its regulation of host-microbe interactions. Gut Microbes 15, 2190300 (2023).
Nagashima, K., Zhao, A., Atabakhsh, K., Bae, M., Blum, J. E. Weakley, Allison. et al. Mapping the T cell repertoire to a complex gut bacterial community. Nature. 621, 162–170 (2023).
Guthrie, G. D., Nicholson-Guthrie, C. S. & Leary, H. L. Jr. A bacterial high-affinity GABA binding protein: isolation and characterization. Biochem. Biophys. Res. Commun. 268, 65–68 (2000).
Kanangat, S., Bronze, M. S., Meduri, G. U., Postlethwaite, A., Stentz, F. et al. Enhanced Extracellular Growth of Staphylococcus aureus in the Presence of Selected Linear Peptide Fragments of Human Interleukin (IL)-1 β and IL-1 Receptor Antagonist. J. infect. dis. 183, 65–69 (2001).
McCracken, V. J., Chun, T., Baldeón, M. E., Ahrné, S., Molin, G. et al. TNF-à Sensitizes HT-29 Colonic Epithelial Cells to Intestinal Lactobacilli1. Exp .biol. med. 227, 665–670 (2002).
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P. & Kaper, J. B. Bacteria–host communication: the language of hormones. Proc. Natl Acad. Sci. 100, 8951–8956 (2003).
Bashir, M. E. H., Louie, S., Shi, H. N. & Nagler-Anderson, C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 172, 6978–6987 (2004).
Wu, L. et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science 309, 774–777 (2005).
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C. & Sperandio, V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl Acad. Sci. 103, 10420–10425 (2006).
Hegde, M., Wood, T. K. & Jayaraman, A. The neuroendocrine hormone norepinephrine increases Pseudomonas aeruginosa PA14 virulence through the las quorum-sensing pathway. Appl. Microbiol. Biotechnol. 84, 763–776 (2009).
Stevens, M. P. Modulation of the interaction of enteric bacteria with intestinal mucosa by stress-related catecholamines. Microbial Endocrinol. 874, 111–134 (2010).
Blier, A.-S. et al. C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology 157, 1929 (2011).
Gruber, C. & Sperandio, V. Regulation in Response to Host-Derived Signaling Molecules. Regul. Bacterial Virulence, 545–565 (2012).
Paino, A. et al. Identification of a novel bacterial outer membrane interleukin-1Beta-binding protein from Aggregatibacter actinomycetemcomitans. PLoS One 8, e70509 (2013).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672 (2014).
Anisa, S., Ismail, J. S. V. & Bassler, B. L. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe 19, 470–480 (2016).
Wang, H. et al. An essential bifunctional enzyme in Mycobacterium tuberculosis for itaconate dissimilation and leucine catabolism. Proc. Natl Acad. Sci. USA 116, 15907–15913 (2019).
Riquelme, S. A. et al. Pseudomonas aeruginosa Utilizes Host-Derived Itaconate to Redirect Its Metabolism to Promote Biofilm Formation. Cell Metab. 31, 1091–1106.e1096 (2020).
Rosenberg, G. et al. Host succinate is an activation signal for Salmonella virulence during intracellular infection. Science 371, 400–405 (2021).
Ahmed, M. et al. Mycobacterium tuberculosis senses host Interferon-gamma via the membrane protein MmpL10. Commun. Biol. 5, 1317 (2022).
Liu, B. et al. Enterohaemorrhagic E. coli utilizes host- and microbiota-derived L-malate as a signaling molecule for intestinal colonization. Nat. Commun. 14, 7227 (2023).
Aleti, G., Troyer, E. A. & Hong, S. G protein-coupled receptors: A target for microbial metabolites and a mechanistic link to microbiome-immune-brain interactions. Brain Behav. Immun. Health 32, 100671 (2023).
Wang, G. et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell. Mol. Life Sci. CMLS 76, 3917–3937 (2019).
Donia, M. S. & Fischbach, M. A. Small molecules from the human microbiota. Science 349, 1254766 (2015).
Liu, Y., Hou, Y., Wang, G., Zheng, X. & Hao, H. Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host-Microbe Interplay. Trends Endocrinol. Metab. 31, 818–834 (2020).
Lokau, J. & Garbers, C. Biological functions and therapeutic opportunities of soluble cytokine receptors. Cytokine Growth Factor Rev. 55, 94–108 (2020).
Wu, X. et al. Gpr35 shapes gut microbial ecology to modulate hepatic steatosis. Pharmacol. Res. 189, 106690 (2023).
Jing, W. et al. Berberine improves colitis by triggering AhR activation by microbial tryptophan catabolites. Pharmacol. Res. 164, 105358 (2021).
Kaur, H., Ali, S. A. & Yan, F. Interactions between the gut microbiota-derived functional factors and intestinal epithelial cells – implication in the microbiota-host mutualism. Front. Immunol. 13, 1006081 (2022).
Neuman, H., Debelius, J. W., Knight, R. & Koren, O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 39, 509–521 (2015).
Kendall, M. M. & Sperandio, V. What a Dinner Party! Mechanisms and Functions of Interkingdom Signaling in Host-Pathogen Associations. mBio 7, e01748 (2016).
Knecht, L. D. et al. Serotonin Activates Bacterial Quorum Sensing and Enhances the Virulence of Pseudomonas aeruginosa in the Host. Ebiomedicine, 9, 161–169 (2016).
Nogal, A., Valdes, A. M. & Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 13, 1–24 (2021).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016).
Husted, A. S., Trauelsen, M., Rudenko, O., Hjorth, S. A. & Schwartz, T. W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 25, 777–796 (2017).
Aisenberg, W. H. et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci. Rep. 6, 38231 (2016).
Thibaut, M. M. & Bindels, L. B. Crosstalk between bile acid-activated receptors and microbiome in entero-hepatic inflammation. Trends Mol. Med. 28, 223–236 (2022).
Studer, E. et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 55, 267–276 (2012).
Keitel, V., Stindt, J. & Häussinger, D. Bile acid-activated receptors: GPBAR1 (TGR5) and other G protein-coupled receptors. Handb. Exp. Pharmacol. 256, 19–49 (2019).
Staudinger, J. L. et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl Acad. Sci. 98, 3369–3374 (2001).
Moore, L. B. et al. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 16, 977–986 (2002).
Chen, M. L. et al. CAR directs T cell adaptation to bile acids in the small intestine. Nature 593, 147–151 (2021).
Makishima, M. et al. Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313–1316 (2002).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Roager, H. M. & Licht, T. R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 9, 3294 (2018).
Ji, J. & Qu, H. Cross-regulatory Circuit Between AHR and Microbiota. Curr. Drug Metab. 20, 4–8 (2019).
Kumar, P., Lee, J. H. & Lee, J. Diverse roles of microbial indole compounds in eukaryotic systems. Biol. Rev. Camb. Philos. Soc. 96, 2522–2545 (2021).
Kumar, A. & Sperandio, V. Indole Signaling at the Host-Microbiota-Pathogen Interface. mBio. 10, e01031–19 (2019).
Zsila, F., Ricci, M., Szigyarto, I. C., Singh, P. & Beke-Somfai, T. Quorum Sensing Pseudomonas Quinolone Signal Forms Chiral Supramolecular Assemblies With the Host Defense Peptide LL-37. Front. Mol. Biosci. 8, 742023 (2021).
Antonioli, L., Blandizzi, C., Pacher, P., Guilliams, M. & Haskó, G. Quorum sensing in the immune system. Nat. Rev. Immunol. 18, 537–538 (2018).
Chen, H. et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 177, 1217–1231.e1218 (2019).
Jin, L. et al. MPYS Is Required for IFN Response Factor 3 Activation and Type I IFN Production in the Response of Cultured Phagocytes to Bacterial Second Messengers Cyclic-di-AMP and Cyclic-di-GMP. J. Immunol. 187, 2595–2601 (2011).
Yin, Q. et al. Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING. Mol. Cell 46, 735–745 (2012).
Salazar, F. & Brown, G. D. Antifungal innate immunity: a perspective from the last 10 years. J. Innate Immun. 10, 373–397 (2018).
Dang, E. V. et al. Secreted fungal virulence effector triggers allergic inflammation via TLR4. Nature 608, 161–167 (2022).
Leal Rodriguez, C. et al. The infant gut virome is associated with preschool asthma risk independently of bacteria. Nat. Med. 30, 138–148 (2024).
Seo, S. U. & Kweon, M. N. Virome-host interactions in intestinal health and disease. Curr. Opin. Virol. 37, 63–71 (2019).
Wahida, A., Tang, F. & Barr, J. J. Rethinking phage-bacteria-eukaryotic relationships and their influence on human health. Cell Host Microbe 29, 681–688 (2021).
Lesouhaitier, O. et al. Gram-negative bacterial sensors for eukaryotic signal molecules. Sensors 9, 6967–6990 (2009).
Gao, D. et al. The roles of cell-cell and organ-organ crosstalk in the type 2 diabetes mellitus associated inflammatory microenvironment. Cytokine Growth Factor Rev. 66, 15–25 (2022).
Heinken, A. & Thiele, I. Systems biology of host-microbe metabolomics. Wiley Interdiscip. Rev. Syst. Biol. Med 7, 195–219 (2015).
Lyte, M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 12, 14–20 (2004).
Lyte, M. & Ernst, S. Catecholamine induced growth of gram negative bacteria. Life Sci. 50, 203–212 (1992).
Luqman, A., Kharisma, V. D., Ruiz, R. A. & Gotz, F. In Silico and in Vitro Study of Trace Amines (TA) and Dopamine (DOP) Interaction with Human Alpha 1-Adrenergic Receptor and the Bacterial Adrenergic Receptor QseC. Cell Physiol. Biochem 54, 888–898 (2020).
Ozuna, H., Uriarte, S. M. & Demuth, D. R. The Hunger Games: Aggregatibacter actinomycetemcomitans Exploits Human Neutrophils As an Epinephrine Source for Survival. Front. Immunol. 12, 707096 (2021).
Zakharevich, N. V. et al. Complete Genome Sequence of Bifidobacterium angulatum GT102: Potential Genes and Systems of Communication with Host. Russian J. Genet. 55, 847–864 (2019).
Hu, S. et al. Gut microbial beta-glucuronidase: a vital regulator in female estrogen metabolism. Gut Microbes 15, 2236749 (2023).
Al-Zawity, J. et al. Effects of the Sex Steroid Hormone Estradiol on Biofilm Growth of Cystic Fibrosis Pseudomonas aeruginosa Isolates. Front. Cell Infect. Microbiol. 12, 941014 (2022).
Kreimendahl, S. & Pernas, L. Metabolic immunity against microbes. Trends Cell Biol. 34, 496–508 (2024).
Messina, J. M. et al. Unveiling cytokine charge disparity as a potential mechanism for immune regulation. Cytokine Growth Factor Rev. 77, 1–14 (2024).
Xu, S., Wang, Q. & Ma, W. Cytokines and soluble mediators as architects of tumor microenvironment reprogramming in cancer therapy. Cytokine Growth Factor Rev. 76, 12–21 (2024).
Porat, R., Clark, B. D., Wolff, S. M. & Dinarello, C. A. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science 254, 430–432 (1991).
McLaughlin, R. A. & Hoogewerf, A. J. Interleukin-1beta-induced growth enhancement of Staphylococcus aureus occurs in biofilm but not planktonic cultures. Micro. Pathog. 41, 67–79 (2006).
Ahlstrand, T., Kovesjoki, L., Maula, T., Oscarsson, J. & Ihalin, R. Aggregatibacter actinomycetemcomitans LPS binds human interleukin-8. J. Oral. Microbiol. 11, 1549931 (2019).
Han, B. et al. Tsr chemoreceptor interacts with IL-8 provoking E. coli transmigration across human lung epithelial cells. Sci. Rep. 6, 31087 (2016).
Mahdavi, J. et al. Pro-inflammatory cytokines can act as intracellular modulators of commensal bacterial virulence. Open Biol. 3, 130048 (2013).
Lee, J. H. et al. Modulation of bacterial growth by tumor necrosis factor-alpha in vitro and in vivo. Am. J. Respir. Crit. Care Med. 168, 1462–1470 (2003).
Luo, G., Niesel, D. W., Shaban, R. A., Grimm, E. A. & Klimpel, G. R. Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun. 61, 830–835 (1993).
Högbom, M. & Ihalin, R. Functional and structural characteristics of bacterial proteins that bind host cytokines. Virulence 8, 1592–1601 (2017).
Zhao, Y. et al. Virulence factor cytotoxin-associated gene A in Helicobacter pylori is downregulated by interferon-γ in vitro. FEMS Immunol. Med. Microbiol. 61, 76–83 (2011).
Tan, L. et al. Engineered probiotics biofilm enhances osseointegration via immunoregulation and anti-infection. Sci. Adv. 6, eaba5723 (2020).
Sigurlásdóttir, S. et al. Host cell-derived lactate functions as an effector molecule in Neisseria meningitidis microcolony dispersal. PLOS Pathog. 13, e1006251 (2017).
Hooftman, A. & O’Neill, L. A. J. The Immunomodulatory Potential of the Metabolite Itaconate. Trends Immunol. 40, 687–698 (2019).
Tomlinson, K. L. et al. Staphylococcus aureus induces an itaconate-dominated immunometabolic response that drives biofilm formation. Nat. Commun. 12, 1399 (2021).
Schubert, C. & Unden, G. C4-dicarboxylates as growth substrates and signaling molecules for commensal and pathogenic enteric bacteria in mammalian intestine. J. bacteriol. 204, e00545–21 (2022).
Nguyen, B. D. et al. Import of Aspartate and Malate by DcuABC Drives H(2)/Fumarate Respiration to Promote Initial Salmonella Gut-Lumen Colonization in Mice. Cell Host Microbe 27, 922–936.e926 (2020).
Schubert, C. et al. C4-dicarboxylates and l-aspartate utilization by Escherichia coli K-12 in the mouse intestine: l-aspartate as a major substrate for fumarate respiration and as a nitrogen source. Environ. Microbiol. 23, 2564–2577 (2021).
O’Boyle, N., Connolly, J. P. R., Tucker, N. P. & Roe, A. J. Genomic plasticity of pathogenic Escherichia coli mediates d-serine tolerance via multiple adaptive mechanisms. Proc. Natl Acad. Sci. 117, 22484–22493 (2020).
Fernandez, M. et al. Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins. Int. J. Mol. Sci. 20, 5156 (2019).
Reyes-Darias, J. A. et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 97, 488–501 (2015).
Rico-Jiménez, M. et al. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol. Microbiol. 88, 1230–1243 (2013).
Planamente, S. et al. Structural basis for selective GABA binding in bacterial pathogens. Mol. Microbiol. 86, 1085–1099 (2012).
Montaudouin, C. et al. Quorum Sensing Contributes to Activated IgM-Secreting B Cell Homeostasis. J. Immunol. 190, 106–114 (2013).
Bardou, M. et al. Quorum sensing governs collective dendritic cell activation in vivo. EMBO J. 40, e107176 (2021).
Tizzano, M. & Sbarbati, A. Hormone fatty acid modifications: Gram negative bacteria and vertebrates demonstrate common structure and function. Med. Hypotheses 67, 513–516 (2006).
Heaver, S. L., Johnson, E. L. & Ley, R. E. Sphingolipids in host-microbial interactions. Curr. Opin. Microbiol. 43, 92–99 (2018).
Brown, E. M. et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 25, 668–680.e667 (2019).
Schroeder, B. O. & Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089 (2016).
Mayer, E. A., Nance, K. & Chen, S. The gut–brain axis. Annu. Rev. Med. 73, 439–453 (2022).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Albillos, A., De Gottardi, A. & Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 72, 558–577 (2020).
Qiu, S. et al. Small molecule metabolites: discovery of biomarkers and therapeutic targets. Sig. Transduct. Target. Ther. 8, 132 (2023).
Rinschen, M. M., Ivanisevic, J., Giera, M. & Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 20, 353–367 (2019).
Loke, P. & Harris, N. L. Networking between helminths, microbes, and mammals. Cell Host Microbe 31, 464–471 (2023).
Tan, J. K., McKenzie, C., Mariño, E., Macia, L. & Mackay, C. R. Metabolite-Sensing G Protein–Coupled Receptors—Facilitators of Diet-Related Immune Regulation. Annu. Rev. Immunol. 35, 371–402 (2017).
Kasubuchi, M., Hasegawa, S., Hiramatsu, T., Ichimura, A. & Kimura, I. Dietary Gut Microbial Metabolites, Short-chain Fatty Acids, and Host Metabolic Regulation. Nutrients 7, 2839–2849 (2015).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32300022), National Key Research and Development Program of China (No. 2019YFA0905600, 2020YFA0907900), and the Funds for Creative Research Groups of China (21621004).
Author information
Authors and Affiliations
Contributions
S.B.W. and X.Y.B. are co-responsible for the collection, collation, and writing of the original manuscript. D.L.C. designed and revised the manuscript. X.Y.W., J.J.Q., H.W., and Q.C. are responsible for the concept development, revision, and review of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, S., Bu, X., Chen, D. et al. Molecules-mediated bidirectional interactions between microbes and human cells. npj Biofilms Microbiomes 11, 38 (2025). https://doi.org/10.1038/s41522-025-00657-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00657-2
This article is cited by
-
MitoCommun: a database for decoding mitochondrial communication networks
BMC Genomics (2026)