Abstract
Migratory birds carry pathogens, posing a significant threat to environmental and human health. We documented the metatranscriptome and RNA virome of 896 stool samples from migratory birds and environmental samples over four consecutive years in southwest China. Our analysis identified Catellicoccus marimammalium as the predominant bacterium in the gut of black-headed gulls, with an average relative abundance of 79.3%. Strain-level analysis of C. marimammalium revealed a dominant population with some longitudinal diversity over the four years. Additionally, the gut of black-headed gulls was found to harbor numerous viruses, including a novel hepatovirus. Lysates of cells of C. marimammalium but not other bacteria derived from black-headed gulls could inhibit the replication of human hepatovirus, suggesting a potential regulatory role for gut commensal bacteria in modulating viral carriage. These findings enhance our understanding of the microbiome and RNA virome diversity in migratory birds and provide insights into the modulation of asymptomatic infections.
Similar content being viewed by others
Introduction
Migratory birds can harbor both viruses and bacteria, some of which may pose a threat to environmental and human health1,2. The long-distance migration by these birds further amplifies this public health risk, as it exposes them to diverse ecosystems and populations3. Migratory birds are natural reservoirs of various viruses, such as avian influenza virus, foot and mouth disease virus, and Japanese encephalitis virus4,5,6. Since the 2020 outbreak of SARS-CoV-2, there has been increased focus on zoonotic infections in wild animals, especially migratory birds. By monitoring wildlife for disease indicators, scientists can potentially identify reservoirs and implement preventive measures to mitigate outbreaks7. Besides viruses, migratory birds can harbor a considerable abundance of pathogenic bacteria, including Escherichia coli, Salmonella, and Listeria monocytogenes8. Several studies indicate that migratory birds also acquire antibiotic-resistant bacteria from polluted surroundings in regions with elevated livestock or human concentrations, and subsequently disseminate these bacteria along their migration routes9. Identifying existing or potential pathogens within these species can help trace the origins of specific epidemics and provide a risk assessment of the most likely sources of future outbreaks.
Black-headed gull (Larus ridibundus) is one of the most prevalent migratory bird species in China. They migrate from Siberia to the southwest of China for wintering every year from November to March10. The black-headed gull has a wide distribution and is known to carry both pathogenic bacteria and viruses8,11. These gulls are often attracted to easily accessible food associated with human activities and are also frequently observed in the vicinity of poultry and livestock farming areas12. To diminish black-headed gull transmission of pathogens to poultry, mammals, and humans, it is valuable to monitor gulls for pathogenic microorganisms.
The tendency of birds to serve as viral reservoirs and asymptomatic carriers is associated with their living habits, including high population densities and close social interactions. Despite these observations, there is a limited understanding of the underlying mechanisms13. It has been suggested that the gut microbiome can either assist the host in defending against viral infestation by triggering immune responses or facilitate the virus in evading the host’s defense through direct and indirect mechanisms14. For example, the gut microbiome of mammals can regulate enteroviruses by modulating the adhesion and colonization of viruses15,16. Thus, an in-depth exploration of the interplay between commensal bacteria and viruses in birds could offer insights into microbiome modulation of viral carriage.
In this study, we investigated the microbiome and virome of migratory birds, primarily black-headed gulls, which migrated to Sichuan Province, China, over the period from 2020 to 2023. We found that the bacterium C. marimammalium (Order Lactobacillales) was dominant in the stool of black-headed gulls, and analysis at the strain level over four years revealed a diverse population. The genome of a highly abundant hepatovirus-like virus was also recovered. Further in vitro experiments indicated that C. marimammalium can regulate the replication of the human hepatovirus, suggesting a potential role of the gut microbiome in the modulation of the viral carriage in these migratory gulls.
Results
C. marimammalium predominates in the stool microbiome of black-headed gulls
To conduct a comprehensive microbial analysis of the black-headed gull, we established research teams in four distinct geographical regions (Deyang, Leshan, Hanyuan, and Qionghai) located in Sichuan Province, China. Over 4 consecutive years, 2020 to 2023, we collected a total of 624 stool samples from black-headed gulls, 124 stool samples from other bird species, and 148 environmental samples (Fig. 1a and Supplementary Table 1). Collection teams distinguished the fecal samples from various bird species, while the types of bird food were identified based on the color variation in their stools (Supplementary Fig. 1A).
a Geographic locations of sample collection sites, with sample size from each region. The migratory bird species featured in this study. They were photographed in their native environments by the sampling teams. Upper left and right: black-headed gull; left center: common coot; bottom left: great cormorant; right center: ruddy shelduck; bottom right: gadwall. Samples were collected in November 2020, December 2021, November and December 2022, as well as February, November, and December 2023. b The taxonomic classification of all the samples at the species level (>40% in at least one sample). The Y axis represents each sample. The right is the occurrence frequency of bacterial species in different sample types. c GO term annotation of genes derived from different bacterial species. The heat map color represents the abundance of each GO term contributed by each bacterium. The pie charts represent the contribution rate of Catellicoccus marimammalium (dark plate) in all functional terms among different sample types. d Resistome analysis of microbiome data. The Y axis represents the top selected antibiotic-resistant genes, and the circle size represents the frequency of genes detected in black-headed gulls and the environment. e Volcano diagram of differentially expressed Catellicoccus marimammalium genes in vivo versus in vitro. The dots in red represent the significantly differentially expressed genes (FDR < 0.05, FoldChange > 2, or FoldChange < 0.5).
Samples were grouped by species, location, and diet, with every four from the same category pooled for RNA extraction and library construction (n = 224). The bacterial and archeal taxonomic analysis revealed a dominant bacterial species in the stool samples of black-headed gull (Fig. 1b). The average abundance of Catellicoccus marimammalium in black-headed gulls was 79.3% ± 2.9% (mean ± s.e.m.), significantly exceeding the levels observed in other birds and environmental samples (Supplementary Fig. 1B). This suggested a distinctive microbial community present in the intestinal tract of black-headed gulls. C. marimammalium could be detected in 96% of all black-headed gull samples, and other commonly identified microbes included Saccharomyces cerevisiae, E. coli, and Clostridium perfringens (Fig. 1b). C. perfringens was the only microbe detected in black-headed gulls exclusively (Fig. 1b). No significant differences in the total microbial community were observed associated with geography or diet (Supplementary Fig. 1B).
The functional landscape, resistome, and virulence factors of C. marimammalium
To explore the potential role of the gut microbiome in black-headed gulls, we profiled the functional activity of microbial genes using metatranscriptomic data. A total of 14,092 GO terms were identified. C. marimammalium contributed the majority of the top functions, such as glycolytic process, DNA replication, and ATP synthesis coupled proton transport (Fig. 1c and Supplementary Fig. 1C). The top 10 pathways in the black-headed gull microbiome differed from other sample types. For instance, genes involved in the glycolytic process were more abundant in the gull microbiome compared to other groups (Supplementary Fig. 1D).
The resistome was analyzed by mapping metagenomic reads to the Comprehensive Antibiotic Resistance Database17. A total of 67 terms were identified. The most abundant was lnuC, which is a transposon-mediated nucleotidyltransferase that confers resistance to lincomycin (Supplementary Fig. 2A). Only two antibiotic-resistant genes, pp_cat and fexA, were shared between black-headed gulls and the environment. These genes were not dominant in black-headed gulls, indicating limited co-occurrence of antibiotic-resistant genes between the birds and the environment. (Fig. 1d).
To assess the transcriptomic profile of C. marimammalium, we isolated 16 bacterial strains from stool samples (Supplementary Table 2). C. marimammalium is a Gram-positive bacterium (Supplementary Fig. 1E). The complete genome of this bacterium underwent sequencing through long-read techniques, and its genomic characteristics were annotated. The genome of C. marimammalium was 1.2 M in length, and 1924 open reading frames (ORFs) were predicted (Fig. 2d). We aligned metatranscriptomic reads to the annotated genome of C. marimammalium to gain insight into the in vivo transcriptome. No transcriptomic differences were observed associated with geography or diet (Supplementary Fig. 2B). We then conducted a comparison between the transcriptome of C. marimammalium in vitro culture and within the gut environment, revealing differential expression in 30 genes (Fig. 1e). These genes were mainly associated with the CRISPR prokaryotic immune defense system and tyrosine-tRNA ligase, suggesting that the gut environment can alter the transcriptome of this bacterium.
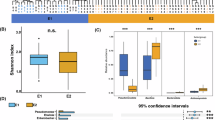
a The VFDB categories of the identified virulent factors in the genome of Catellicoccus marimammalium. b The gene expression of potential virulence factors in Catellicoccus marimammalium. The density chart shows the relative expression pattern of each VFDB term across all samples. The X axis represents the relative expression level of different terms. c Strain-level analysis of Catellicoccus marimammalium. ANI values (x axis) obtained from InStrain of all black-headed gull samples. Colors are assigned based on 99.9% ANI. Strains considered to be the same strain will be annotated with the same color. Six main clades are shown in different colors in the phylogenetic tree. Other metadata is indicated using cells of different colors. HY Hanyuan, LS Leshan, QH Qionghai, DY Deyang. d The genome annotation of Catellicoccus marimammalium. e The alteration of Catellicoccus marimammalium clades in black-headed gulls upon arrival and departure. f Distribution patterns of six main Catellicoccus marimammalium clades across different years.
We next analyzed the virulence factor(s) of C. marimammalium by mapping the predicted ORFs to the virulence factor database (VFDB)17. In total, 168 terms were identified, belonging to 13 categories (Fig. 2a). Prominently expressed virulence factors included lipoteichoic acid biosynthesis proteins and collagen adhesins. The sequencing data were then mapped to these VF regions, and most of them were actively expressed both in vivo and in vitro, suggesting the potential pathogenicity of this bacterium (Fig. 2b).
Strain-level tracking of C. marimammalium in the stool of black-headed gulls
The strain-level analysis was conducted among all black-headed gull samples to track the longitudinal and geographical dynamics of C. marimammalium. This analysis identified six clades of C. marimammalium, with no consistent bias associated with geography or year of collection (Fig. 2c, Supplementary Fig. 3A, D). The six clades exhibited varying distribution patterns across different years. Clade 3 remained dominant in black-headed gull samples from 2020 to 2023 (Fig. 2f). The diversity of C. marimammalium fluctuated over the years, with the lowest diversity observed in 2021 and the highest in 2022 (Supplementary Fig. 3B).
To understand the impact of the local environment on the strain-level distribution of C. marimammalium, we compared the stool microbiomes of black-headed gulls immediately after their arrival and before their departure from Sichuan in 2022 and 2023. Despite Clade 3 remaining the predominant subtype of C. marimammalium, its relative abundance notably increased before its departure. Additionally, Clade 5 was present upon their arrival but disappeared after several months of residence, while Clade 2 emerged during their stay. These findings underscore the local strain-level dynamics of C. marimammalium (Fig. 2e and Supplementary Fig. 3C). To be noted, the sampling bias may affect some of the conclusions, especially for the strain-level analysis. Future studies with larger sample sizes and more comprehensive sampling strategies could further validate our findings.
The RNA virome profiling revealed a hepatovirus-like virus dominant in the stool of the black-headed gull
RNA viruses could be characterized in the metatranscriptomic data from the stool of black-headed gulls. A total of 56 virus families were identified; 24 viral families were identified in at least five samples (Fig. 3a). Those viruses included vertebrate viruses, invertebrate viruses, plant viruses, and others, revealing a high diversity of the RNA virome (Fig. 3b). More viruses that can infect vertebrates were identified in birds when compared to the environmental samples (Fig. 3c, d). The major viral families identified were Picornaviridae, Sedoreoviridae, and Caliciviridae (Fig. 3e). None of these samples contained detectable viruses closely related to SARS-CoV-2 or influenza viruses.
a The overview of the RNA virome. The top heat map represents the family of RNA viruses that can infect vertebrates, and the bottom heatmap represents the family of RNA viruses that infect non-vertebrates. The heat map color represents the relative abundance of different viral families. Other metadata is indicated using cells of different colors. The viral families that can be identified in at least five samples are shown in this heat map. b The host of the identified viruses in each sample. The X axis represents the different samples, and the Y axis represents the relative abundance of different groups. The number of non-vertebrate viruses (c) and vertebrate viruses (d) identified in different sample types. e The taxonomic classification of identified vertebrate viruses at the family level. The X axis represents the number of samples containing the corresponding viruses, and the Y axis represents the number of viral species belonging to the corresponding families.
The majority of the viral contigs of Picornaviridae belonged to the Hepatovirus genus (Fig. 4a). We then selected all the reads that could be mapped to this genus and assembled them into 87 representative viral genomes. A total of 16 abundant viral genomes were selected for further analysis (Fig. 4b). Phylogenetic analysis of the representative genomes revealed that they were related to the avian hepatovirus (Fig. 4c). Genomic annotation indicated the presence of picornavirus/calicivirus coat-like protein, Hepatitis A virus-like VP protein, and viral RNA-dependent RNA polymerase, as expected for the genus Hepatovirus (Fig. 4d).
a The taxonomic classification of identified viral contigs belonging to the Picornaviridae family. b The count of reads that can be aligned with the contigs is associated with the hepatovirus-like virus. The heat map color represents the read number of the viral contigs in different samples. The X axis represents different samples, and the Y axis represents different viral contigs. c Phylogenetic analysis of the selected novel hepatovirus-like virus RdRP sequences. d The genome annotation for the selected novel hepatovirus-like virus genome.
Cell lysate of C. marimammalium inhibits the growth of the human hepatovirus
Bacteria can regulate the infection of viruses14. To identify the possible influence of the bacterial community in black-headed gulls on viral abundance, we analyzed the association between the abundance of bacteria and RNA viruses. The results indicated that the abundance of hepatovirus-like viruses was highly associated with the C. marimammalium (Fig. 5b) such that the two showed a strong negative correlation (Fig. 5a). Therefore, we hypothesized that C. marimammalium may inhibit the infection of the hepatovirus-like virus.
a The correlation analysis on the abundance of Catellicoccus marimammalium and the hepatovirus-like virus (Spearman correlation). b Random forest analysis to evaluate the association between the abundance of Catellicoccus marimammalium and different viral genera. The X axis represents the mean decrease in accuracy value and the mean decrease in the Gini coefficient, and the higher the value of these two scores, the higher the importance of the variable in the model. c The experimental design for the evaluation of the effects of Catellicoccus marimammalium on the infectivity of HM175-18f. d The inhibitory effects of Catellicoccus marimammalium lysate on the replication of HM175-18f. The Y axis represents the RNA level of the viruses, and the X axis represents the time after co-culture. The viral RNA was tested in both cells and culture supernatant. e The inhibitory effects of different bacteria isolated from the stool of black-headed gull on the replication of HM175-18f. The Y axis represents the RNA level of the viruses, and the X axis represents different samples. The data shown in the plot represent the samples collected 72 hours after co-culture. The viral RNA was tested in both cells and culture supernatant.
We used an infectious clone for HM175-18f, a human hepatovirus A strain commonly used as an in vitro model, to test the viral infectivity after co-culture with C. marimammalium. HM175-18f viruses were used to infect Huh7.5.1 cells, and after a 5-hour incubation period, bacterial cell lysate or bacterial cell secretion products were introduced into the culture medium (Fig. 5c). We found that the bacterial cell lysate, but not the bacterial cell secretion products, could inhibit the growth of HM175-18f more than 1000-fold following 48 hours of co-culture (Fig. 5d, e). Other bacteria that were isolated from the stool of black-headed gulls could inhibit the growth of HM175-18f (Fig. 5e). With the reduced titration of C. marimammalium cell lysate, a corresponding decrease in the amount of HM175-18f viruses was observed. This provided additional confirmation of the inhibitory role of the bacterial cell lysate (Supplementary Fig. 4A). We then conducted an experiment involving the addition of bacterial cell lysate at the initiation of the experiment, followed by its removal after a 5-hour co-culture period (Supplementary Fig. 5A). Notably, no inhibitory effect on virus growth was discerned over the entire 96-hour duration, suggesting that the inhibitory process primarily occurred after the virus had entered the cell, as opposed to during the pre-adsorption process (Supplementary Fig. 5B, C). The cytotoxicity of the C. marimammalium cell lysate in Huh7.5.1 cells was also evaluated. The cell lysate exhibited no significant cytotoxicity at the concentration that inhibited the viral growth significantly, suggesting that the bacterial lysate could not reduce cell viability at the concentrations tested (Supplementary Fig. 5D).
Discussion
This study characterized the bacterial and viral communities present in stool samples of a population of migratory birds, incorporating metagenomic data of the full community and longitudinal data from metatranscriptomic analysis. The findings provide valuable insights into both the taxonomic and functional aspects of these microbial communities and specify a potential regulatory role of bacteria in modulating the abundance of a viral pathogen in the bird gut.
Birds belonging to the Laridae family appear to harbor a substantial quantity of C. marimammalium18,19. The presence of C. marimammalium 16S DNA has been documented previously, constituting ~55% of the seagull stool microbiome19. Our results reveal an exceptionally high level of colonization and gene expression for C. marimammalium within the gastrointestinal tract of black-headed gulls. The microbiome of black-headed gulls differs from that of other birds in the same area, suggesting that host genetics may have a more significant role than environmental influences in determining gut microbial colonization. This aligns with previous findings20,21. The black-headed gull demonstrates a high degree of adaptability to external environments22, and we attempt to address this high adaptability from the perspective of the gut microbiome. The intestinal microbiome of most mammals and higher animals, including birds, generally lacks a single species with overwhelming dominance23. Bacteria with overwhelming dominance are more commonly found in the intestinal microbiome of insects or certain fish species23. It has been reported that external factors, including diet, environment, geography, and others, can induce alterations in both the taxonomic and functional aspects of the vertebrate fecal microbiome24,25,26,27,28. C. marimammalium remains notably stable, showing resilience to alterations in its surroundings. The possible impact of a single dominant gut bacterial species on the functions of black-headed gulls is intriguing. A more in-depth examination is necessary to fully understand the functional dynamics. To achieve this, metabolomic analysis can be employed to provide a more comprehensive perspective on the functionality of these bacteria in response to varying environmental conditions.
We performed a strain-level analysis on Catellicoccus marimammaliu, which allowed us to characterize genetic and phenotypic variations. It has been reported that microbial strain variants can affect the physiology of the microbial community and the host29,30,31. Furthermore, migration patterns are associated with the avian microbiome composition and diversity32,33. In this study, we observed a longitudinal pattern in the strain-level dynamics of C. marimammalium. Although the primary strain remained consistent across different years and locations, we observed moderate alterations in the strain composition. New strains were observed to emerge during the residence of these birds at single locations. The presence of bacteria is influenced by various factors. Environmental factors (such as seasonal changes and food resources) and host-related factors (such as health status and nutritional level) have potential impacts on the bacterial community. Here, the sampling scope may not be extensive enough to identify all potential influencing factors. Due to the complexity of the metatranscriptomic data, we did not emphasize the evolutionary changes of the bacteria. Instead, we aimed to show that the strain-level of C. marimammalium fluctuated over different time periods, resulting in the observed dynamics. Another limitation is that the SNV identification was carried out using metatranscriptomic data, which may provide limited information compared to metagenome or whole genome sequencing. Going forward, isolating a greater number of C. marimammalium strains will be valuable to help specify the variability and genomic landscape.
The metatranscriptomic analysis unveiled a diverse virome in the stool samples of black-headed gulls. This observation aligns with findings from prior research34. We identified a previously unknown virus with similarities to human hepatovirus (the RdRP sequence showed 41% identity with 88% coverage at the amino acid level). This virus is also closely related to a virus identified in other birds, indicating wide distribution35,36. This discovery underscores the potential risk associated with migratory birds serving as reservoirs for viruses, potentially impacting human health4,35,36,37. Subsequent investigations would be useful to assess the toxicity and zoonotic potential of these viruses.
The gut microbiome may act as a regulator in viral infections, either promoting or inhibiting viral14. This monobacterial dominance in gulls offers an opportunity to investigate interactions between viruses and bacteria. We found that C. marimammalium inhibited the replication of a representative human hepatovirus. However, its role in modulating viral carriage in migratory gulls in vivo remains to be determined, and more in vivo studies are needed. Other bacterial lineages were tested as controls and shown to lack inhibitory activity. Previous studies indicated that bacteria may inhibit viral infection by various mechanisms, such as enhancing the host antiviral responses38,39, destabilizing viruses40, or others41,42. The specific substances in the bacterial lysate of C. marimammalium that inhibited viral replication, and the mechanisms involved, have not yet been identified. Further studies are warranted to delve into the mechanisms through which C. marimammalium modulates the replication of hepatoviruses.
In summary, this study contributes to advancing the understanding of the structure and function of the gut microbiome in black-headed gulls, a prominent migratory bird species in China. We found that the dominant bacterium, C. marimammalium, exhibited several features suggesting its adaptability to diverse environments. We also identified a cluster of human hepatovirus-like viruses in the gut of the black-headed gulls. In vitro experiments demonstrated that cell lysates of C. marimammalium inhibited the growth of a human hepatovirus, underscoring its potential role in suppressing viral replication. These findings enhance our understanding of the competition within avian gut microbiomes and also hold promise for the development of proactive measures against zoonotic diseases, particularly risks associated with migratory bird populations.
Methods
Sample collection
The research teams were strategically established in four distinct geographical regions within Sichuan Province, China, to collect samples. Environmental samples were also obtained from the respective areas. Fecal samples were collected within 5 minutes after the birds excreted new feces to minimize external contamination and ensure sample integrity. Sampling activities spanned four consecutive years, covering the period from 2020 to 2023. The types of bird diets were identified based on color variations observed in the stools (Green-Aquatic plants; Yellow-Artificial feed; White-Fish; Brown-Fruit), providing insights into the dietary habits of the sampled bird species. The differentiation and attribution of fecal samples were carried out by experienced professional ornithologists. Environmental samples included soil samples, water samples, artificially fed bread samples (bread crumbs collected in the human-feeding areas), and aquatic plant samples in the vicinity of black-headed gulls roosting sites. About 100 ml of water collected, 20 g soil, 20 g bread, and 20 g aquatic plant were collected. About one gram of fecal material was carefully gathered from the upper layer to prevent any environmental impurities, using a disposable sterile plastic spatula. The samples were then transferred into a 1.5 ml tube with and/or without RNA stabilization buffer. Each tube was promptly placed on dry ice before being stored at -80°C.
Metatranscriptomic sequencing
During the library construction process, four samples were pooled according to the species, location, and diet, for subsequent RNA extraction and library preparation. The prokaryotic RNA library preparation utilizing DNBSEQ technology (BGI, China) was used to prepare libraries. Briefly, total RNA was extracted using custom reagents. The rRNA removal was conducted using Streptavidin magnetic beads based on the specific capture of microbial rRNA sequences(Supplementary Table 3). The first and second-stranded cDNA were then synthesized. The cDNA was subjected to end-repair and adaptor ligation, followed by PCR amplification. The primer sequence was 5ʹ-GAACGACATGGCTACGA-3ʹ (forward) and 5ʹ-TGTGAGCCAAGGAGTTG-3ʹ (reverse).
The taxonomic, functional, and strain-level analysis of the metatranscriptomic data
MetaPhlAn4 (version 4.0; parameters were set as defaults) was used to profile the taxonomic composition of the microbial communities43. Human Project Unified Metabolic Analysis Network 2 (HUMAnN2; human version 3.0.1; parameters were set as defaults)) was employed for functional profiling of the microbiome44. The Ariba (version 2.14.6; parameters were set as defaults) tool and the CARD database were utilized to detect antimicrobial resistance-associated genes45. VF-like genes of C. marimammalium were identified and categorized according to the VFDB database. RPM value was assigned for transcriptional expression of VF-like genes17. The analysis of SNVs in Catellicoccus was conducted using the InStrain(parameters were set as defaults) tool, which analyzes co-occurring genome populations from metagenomes46. The phylogenetic tree was constructed using the InStrain to specifically examine the ANI (Average Nucleotide Identity) among high-quality C. marimammalium in all black-headed gull samples. This tool can determine the nucleotide diversity among samples by mapping sample reads to the representative genome. The default threshold of the software was used.
Isolation of bacterial strains and Gram staining
During the bacterial isolation, five samples with rich microbial diversity and a high probability of containing the target bacteria based on preliminary detection were prioritized. These samples were from different geographical locations, sampling times, and birds with different dietary characteristics to ensure the representativeness of the isolated bacteria. A total of 16 bacterial strains were extracted from the stool samples of black-headed gulls utilizing Brucella Blood Agar plates under micro-anaerobic conditions. The plates were incubated at 37 °C in a 5% CO2 incubator for a maximum of 72 hours. Single colonies were picked and re-cultured in medium plates at least three times to isolate pure bacterial strains and preserved at −80 °C. The bacterial taxonomy was determined by sequencing the full-length 16S rRNA gene (Supplementary Table 2). The primer sequence was 5ʹ-AGAGTTTGATCCTGGCTCAG-3ʹ (forward) and 5ʹ- TACGGCTACCTTGTTACGACTT-3ʹ (reverse). The sequencing results were compared with the database of NCBI using the BLAST tool to determine the taxonomic status of the bacteria(Supplementary Table 2). The relatively small number of 5 fecal samples may not fully represent the vast microbial diversity present in the entire population of migratory birds.
A bacterial culture solution was prepared, and the bacterial smear was fixed onto a slide. The Gram-staining procedure was performed using a Gram stain kit (Solarbio, China) following the manufacturer’s instructions.
Long-read sequencing of bacterial genomes
The DNA of C. marimammalium was extracted using MagMAX™ Viral/Pathogen Ultra Nucleic Acid Isolation Kit (ThermoFisher). The concentration of extracted DNA was measured with a Qubit 4.0 (ThermoFisher Scientific, USA). One library of C. marimammalium was prepared using a Qitan DNA Library prep kit (QDL-E V1.0). Sequencing was performed using a Qcell-3841 sequencing chip and Qitan DNA Sequencing Kit (QDS V1.0) on a QNome platform (QitanTech, China). The library preparation for QitanTech nanopore sequencing was performed, and long-read metagenomic sequencing was performed on the QNome-3841 platform developed by QitanTech (Qitan Technology Co., Beijing, China). The long reads were assembled with Flye47.
RNA virome analysis
For each library, sequencing reads were first quality-controlled using Trimmomatic48. Host reads were moved using Bowtie249. The remaining reads were assembled de novo using MEGAHIT50, deploying default parameters. The assembled contigs of length >1 kb were compared against the NCBI non-redundant protein database (nr) using Diamond blastx51 and the Nucleotide Sequence Database(nt) using blastn52 to achieve the taxonomic assignment. The E-value cut-off was set at 1E-10 to maintain high sensitivity at a low false-positive rate. Taxonomic lineage information was obtained for the top blast hit of each contig, and those classified under the kingdom “Viruses” were identified using taxonomizr (https://github.com/sherrillmix/taxonomizr). Rsem53 (excluding rRNA) was employed to assign a TPM value to each contig to assess the relative abundance. In this study, we employed ICTV information (https://talk.ictvonline.org/) to explore the host range of the identified viruses.
The phylogenetic analysis
Multiple alignments of viral RdRP sequences were performed using MAFFT54. Following sequence alignment, all ambiguously aligned regions were removed using trimAl55. Phylogenetic trees were then estimated for the sequence alignment of each family using the maximum likelihood method available in Iqtree56.
Antiviral activity test
The C. marimammalium culture was placed at 37 °C, LB medium, 5% CO2 incubator overnight and the culture was passed through a 0.22 μm filter to obtain C. marimammalium secretion. The C. marimammalium culture (OD = 0.38, CFU = 2.8 × 104/μl) was craked with Sonic Dismembrators (25 mins), and the C. marimammalium lysate was obtained after passed through a 0.22 μm filter. A concentration step was employed to concentrate the C. marimammalium culture from 20 ml to 1 ml. Other bacteria that were isolated from the stool of black-headed gulls were used the same colony of CFU, craked conditions, and concentration step.
Huh7.5.1 cells were seeded into six-well plates for 24 h. The human hepatovirus infectious clone (HM175-18f) with a multiplicity of infection (MOI) of 0.01 was added to allow infection for 5 h when the cell density was ~20%. After 5 h of infection, the uninfected virus was removed from the cell culture supernatant, and a fresh lysate-containing medium was added. At an interval of 24 hours, medium supernatant and cells were taken separately, and the RNA of HM175-18f was quantified by quantitative real-time RT-PCR. The primer sequence specific for the HM175-18f was 5ʹ-GGTAGGCTACGGGTGAAAC-3ʹ (forward) and 5ʹ-AACAACTCACCAATATCCGC-3ʹ (reverse)57.
The C. marimammalium lysate was added simultaneously with the virus (MOI of 0.01) at time 0 h. After 5h of infection, the virus and C. marimammalium lysate mixture was removed from the cell culture supernatant, and a fresh no-lysate-containing medium was added to verify whether the inhibitory effect occurs before or after the virus entrance. At an interval of 24 hours, medium supernatant and cells were taken separately, and the RNA of HM175-18f was quantified by quantitative real-time RT-PCR.
The C. marimammalium lysate and C. marimammalium secretion components were separated for experimentation to test whether the inhibition effect was specific. Other bacterium strains (Staphylococcus warneri, Streptococcus respiraculi, Escherichia ferguson) isolated from the stool of the black-headed gulls were also selected for lysate suppression experimentation.
Cytotoxicity assay of C. marimammalium lysate
The C. marimammalium culture (OD = 0.38, CFU = 2.8 × 104/μl, volumes = 20 ml) was craked with Sonic Dismembrators (25 mins), C. marimammalium lysate was obtained after being passed through a 0.22 μm filter. The lysate was concentrated to 1 ml with a centrifugal filter. Gradient dilution to obtain different concentrations of lysate. The inhibition rate was quantified by comparing the RNA of HM175-18f with quantitative real-time RT-PCR after adding lysate of different concentrations for incubation.
Cytotoxicity of the C. marimammalium lysate in the cell was tested by using the Cell Counting Kit-8 (CCK-8, Beyotime). Briefly, each cell type was seeded into the wells of a 96-well microtiter plate (10,000 per well) and incubated at 37 °C for 12 h, replacing the medium with C. marimammalium lysate medium containing at graded concentrations to culture at 37 °C for 1 day, CCK-8 solution (10 μL per well) was added, followed by an additional incubation for 0.5 h. The absorbance was measured at 450 nm.
Statistical analysis
All statistical analysis was performed using R and RStudio software. The Wilcoxon rank-sum test was used to compare two independent groups. The Kruskal–Wallis rank-sum test was used for comparing three or more independent groups. Non-parametric correlation was performed using Spearman’s rank-order correlation. The p values for multiple comparisons were corrected using the Benjamini–Hochberg FDR method; p < 0.05 or FDR < 0.05 was considered significant. All acquired data were included in the analyses.
Data availability
The raw sequence data and sample information reported in this paper have been deposited in the Genome Sequence Archive58 in National Genomics Data Center59, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA014153) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Metatranscriptomic data can be accessed at https://ngdc.cncb.ac.cn/gsa/browse/CRA014153. The assembled viral and bacterial genomes/sequences can be accessed at https://ngdc.cncb.ac.cn/genbase/search/gb/C_AA104674.1 and https://ngdc.cncb.ac.cn/biosample/browse/SAMC4694878.
Code availability
All bioinformatic scripts are available on GitHub (https://github.com/luoqingqing777/Migratory-birds).
References
Lycett, S. J. et al. Role for migratory wild birds in the global spread of avian influenza H5N8. Science354, 213–217 (2016).
Hernando-Amado, S., Coque, T. M., Baquero, F. & Martínez, J. L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442 (2019).
Humphreys, J. M. et al. Waterfowl occurrence and residence time as indicators of H5 and H7 avian influenza in North American Poultry. Sci. Rep. 10, 2592 (2020).
Shan, T. et al. Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome 10, 60 (2022).
Keawcharoen, J. et al. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14, 600–607 (2008).
Brown, J. D., Swayne, D. E., Cooper, R. J., Burns, R. E. & Stallknecht, D. E. Persistence of H5 and H7 avian influenza viruses in water. Avian Dis. 51, 285–289 (2007).
He, W.-T. et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 185, 1117–1129.e8 (2022).
Gan, L. et al. Carriage and potential long distance transmission of Listeria monocytogenes by migratory black-headed gulls in Dianchi Lake, Kunming. Emerg. Microbes Infect. 8, 1195–1204 (2019).
Navedo, J. G., Araya, V. & Verdugo, C. Upraising a silent pollution: Antibiotic resistance at coastal environments and transference to long-distance migratory shorebirds. Sci. Total Environ. 777, 146004 (2021).
Chang, H. et al. First report of Chlamydia psittaci seroprevalence in black-headed gulls (Larus ridibundus) at Dianchi Lake, China. Open Life Sci. 13, 250–252 (2018).
Chu, K.-K. et al. Characterization of Deltacoronavirus in Black-Headed Gulls (Chroicocephalus ridibundus) in South China Indicating Frequent Interspecies Transmission of the Virus in Birds. Front. Microbiol. 13, 895741 (2022).
Ferns, P. N. & Mudge, G. P. Abundance, diet and Salmonella contamination of gulls feeding at sewage outfalls. Water Res. 34, 2653–2660 (2000).
Nabi, G. et al. Bats and birds as viral reservoirs: A physiological and ecological perspective. Sci. Total Environ. 754, 142372 (2021).
Li, N., Ma, W.-T., Pang, M., Fan, Q.-L. & Hua, J.-L. The commensal microbiota and viral infection: a comprehensive review. Front Immunol. 10, 1551 (2019).
Kuss, S. K. et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334, 249–252 (2011).
Yin, H.-C. et al. Chicken intestinal microbiota modulation of resistance to nephropathogenic infectious bronchitis virus infection through IFN-I. Microbiome 10, 162 (2022).
Liu, B., Zheng, D., Zhou, S., Chen, L. & Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917 (2022).
Lu, J., Santo Domingo, J. W., Lamendella, R., Edge, T. & Hill, S. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 74, 3969–3976 (2008).
Koskey, A. M., Fisher, J. C., Traudt, M. F., Newton, R. J. & McLellan, S. L. Analysis of the gull fecal microbial community reveals the dominance of Catellicoccus marimammalium in relation to culturable Enterococci. Appl. Environ. Microbiol. 80, 757–765 (2014).
Grond, K., Ryu, H., Baker, A. J., Santo Domingo, J. W. & Buehler, D. M. Gastro-intestinal microbiota of two migratory shorebird species during spring migration staging in Delaware Bay, USA. J. Ornithol. 155, 969–977 (2014).
Ruiz-Rodríguez, M., Martín-Vivaldi, M., Martínez-Bueno, M. & Soler, J. J. Gut microbiota of great spotted cuckoo nestlings is a mixture of those of their foster magpie siblings and of cuckoo adults. Genes9, 381 (2018).
Bonnedahl, J. et al. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum -lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65, 1939–1944 (2010).
Sherrill-Mix, S. et al. Allometry and Ecology of the Bilaterian Gut Microbiome. mBio 9, e00319–18 (2018).
Abu-Ali, G. S. et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol 3, 356–366 (2018).
Wang, D. D. et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343 (2021).
Góngora, E., Elliott, K. H. & Whyte, L. Gut microbiome is affected by inter-sexual and inter-seasonal variation in diet for thick-billed murres (Uria lomvia). Sci. Rep. 11, 1200 (2021).
Grond, K. et al. Composition and drivers of gut microbial communities in arctic-breeding shorebirds. Front. Microbiol. 10, 2258 (2019).
Joakim, R. L. et al. Geography and elevation as drivers of cloacal microbiome assemblages of a passerine bird distributed across Sulawesi, Indonesia. Anim. Microbiome 5, 4 (2023).
Zhang, C. & Zhao, L. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med. 8, 41 (2016).
Yan, Y., Nguyen, L. H., Franzosa, E. A. & Huttenhower, C. Strain-level epidemiology of microbial communities and the human microbiome. Genome Med. 12, 71 (2020).
Park, S.-Y. et al. Strain-level fitness in the gut microbiome is an emergent property of glycans and a single metabolite. Cell 185, 513–529.e21 (2022).
Obrochta, S., Savo Sardaro, M. L., Amato, K. R. & Murray, M. H. Relationships between migration and microbiome composition and diversity in Urban Canada Geese. Front. Ecol. Evol. 10, 742369 (2022).
Thie, N. et al. Linking migration and microbiota at a major stopover site in a long-distance avian migrant. Mov. Ecol. 10, 46 (2022).
Liao, F. et al. Metagenomics of gut microbiome for migratory seagulls in Kunming city revealed the potential public risk to human health. BMC Genomics 24, 269 (2023).
Wille, M., Shi, M., Klaassen, M., Hurt, A. C. & Holmes, E. C. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 13, 2603–2616 (2019).
Wille, M., Shi, M., Hurt, A. C., Klaassen, M. & Holmes, E. C. RNA virome abundance and diversity is associated with host age in a bird species. Virology 561, 98–106 (2021).
Hill, S. C. et al. Impact of host age on viral and bacterial communities in a waterbird population. ISME J. 17, 215–226 (2023).
Steed, A. L. et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502 (2017).
Abt, M. C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Bandoro, C. & Runstadler, J. A. Bacterial lipopolysaccharide destabilizes influenza Viruses. mSphere 2, e00267–17 (2017).
Botic, T., Klingberg, T., Weingartl, H. & Cencic, A. A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int. J. Food Microbiol. 115, 227–234 (2007).
Wang, Z. et al. Inhibitory influence of enterococcus faecium on the propagation of swine influenza A virus in vitro. PLoS One 8, e53043 (2013).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023).
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Hunt, M. et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 3, e000131 (2017).
Olm, M. R. et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat. Biotechnol. 39, 727–736 (2021).
Kolmogorov, M. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Hirai-Yuki, A. et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science353, 1541–1545 (2016).
Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 19, 578–583 (2021).
Xue, Y. et al. Database resources of the national genomics data center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 51, D18–D28 (2023).
Acknowledgements
We are grateful to members of the Liang laboratory for their help and suggestions. We thank Mr. Yanlin Du and Yiping Hu’s contribution to the photography of the birds. We thank Mr. You Shen, Jun Yang, Guoxiang Li, and Yanlin Du for their kind assistance in the collection of samples. This work was supported by the National Forestry and Grassland Administration (2130211). This work was also supported by the Start-up Foundation of Tsinghua University (400-53332101822), Tsinghua University Initiative Scientific Research Program, Tsinghua University Dushi Program (20241080062), Tsinghua-Peking Center for Life Sciences (045-61000100122), and the Natural Science Foundation of China (32200036, 82341116). This work was also supported by the Science & Technology Department of Sichuan Province (2018JY0090).
Author information
Authors and Affiliations
Contributions
Q.L. carried out the biochemical experiments and bioinformatic analysis; H.G. assisted with bioinformatic analysis and artwork; L.W., F.L., and Y.G. carried out sample and metadata collection; Y.X., L.D., J.L., X.W., and C.S. assisted with biochemical manipulations and bioinformatic analysis; Q.D. and C.Q. assisted in data interpretation and draft writing; L.W., Q.L., and G.L. conceived the project and wrote the paper. All authors revised the manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, Q., Gao, H., Xiang, Y. et al. The dynamics of microbiome and virome in migratory birds of southwest China. npj Biofilms Microbiomes 11, 64 (2025). https://doi.org/10.1038/s41522-025-00703-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00703-z