Abstract
The integration of artificial intelligence (AI) in breast cancer pathology has been driven by the promise of “big data”-based foundation models: large deep learning systems deriving diagnostic and prognostic insights from digitized whole slide images (WSIs). Yet, despite progress in computational power and architectures, these models face obvious barriers to clinical use, including poor workflow integration, limited explainability, and reduced generalizability across diverse clinical settings. This article examines the opportunities provided by small, task-oriented AI models designed to predict clinically relevant molecular features in breast cancer, such as hormone receptors (HRs), HER2, Ki-67, BRCA-related status, and somatic mutations directly from WSIs. To overcome foundation model constraints, approaches like model distillation, weak supervision, and modular training are critically examined. Progress now depends on high-quality datasets, rigorous multi-institutional validation, and collaboration between computational scientists, clinicians, and regulators to deliver explainable, clinically actionable innovations in breast cancer.
Similar content being viewed by others
Introduction: the “big data dream” in pathology
Artificial intelligence (AI) is rapidly transforming pathology, offering unprecedented opportunities to improve diagnostic quality and standardization, particularly in complex, high-impact disease areas such as breast cancer1,2. Major drivers of this transformation are the so-called “foundation models”, i.e., self-supervised AI systems trained on large datasets, with pathology whole slide images (WSIs) as a central data source3,4. A key feature of these models is their potential adaptation to diverse downstream tasks (e.g. slide-level classification, region-of-interest analysis, survival prediction, and biomarker discovery)5. Digital pathology enthusiasts and technology companies project these models to perform diagnostic tasks with accuracy comparable to, or even exceeding, that of a human pathologist6,7,8,9. At the same time, novel AI tools are showing fascinating results in predicting molecular alterations directly from WSIs, including breast cancer subtyping and molecular status prediction, suggesting a potential to complement and/or anticipate traditional biomarker testing4,10.
Following the advances in various fields of image recognition (e.g. image search, surveillance, facial/object detection, autonomous driving), the dominant view in digital pathology has been that larger models trained on more data will lead to better performance11,12. This assumption has fueled a renewed emphasis on the “big data” paradigm and the pursuit of increasingly complex model architectures, with the expectation that data scale alone would ensure robustness and generalizability for clinical use13. Notably, this mirrors the early days of genomics, when comprehensive approaches like whole-genome sequencing (WGS) and “multi-omics” were hailed as the ultimate tools for precision oncology14. Yet, over time, the field shifted toward more pragmatic, focused strategies such as targeted sequencing panels, which proved more efficient and clinically actionable15,16,17. In the field of digital pathology, however, the allure of big data has been reborn, raising important questions about whether this approach will ultimately yield similar adjustments toward streamlined, purpose-driven solutions18,19.
While many academic centers and pathology or biobank consortia now generate impressive volumes of data, these resources remain largely underutilized in the development of clinically meaningful and deployable AI solutions20,21,22,23. One of the main barriers is the lack of high-quality data curation, harmonization, and annotation, which are essential, albeit resource-intensive, steps24. As a result, the indiscriminate accumulation of data, especially from digitalized retrospective tissue archives, often results in datasets that are difficult to use, marked by inconsistent metadata and variable quality25. These limitations can lead models not only to overfit, but also to underperform due to label noise, domain shift, or bias, ultimately compromising clinical utility26. Adding to these challenges, the infrastructure required to train and deploy such models is not only computationally intensive and costly, but also environmentally unsustainable27,28. To develop clinically impactful solutions for breast cancer management, the field of computational pathology is called to progressively move toward smarter, task-oriented approaches that prioritize efficiency and scalability.
Limits of large foundation models in breast cancer
In breast cancer, where molecular profiling is essential for diagnosis, prognosis, and therapeutic decision-making, the implementation of foundation models in routine pathology is evolving. Unlike traditional deep learning models trained on narrow, single-purpose datasets, foundation models are designed to exploit extremely large and diverse bodies, often including millions of WSIs from heterogeneous cohorts, precisely to improve generalizability and robustness across tumor types, populations, and institutions29,30. One of the key advantages of foundation models lies in their capacity to act as universal feature extractors, adaptable across multiple downstream tasks and cancer types, rather than requiring bespoke training for each endpoint31. Moreover, it is essential to distinguish between vision-only models and multimodal models that integrate both histological images and textual data32,33. The latter, image-text aligned models, have shown superior generalization capabilities in recent studies, outperforming vision-only approaches in the prediction of clinical biomarkers and subtypes34. For example, a recent work directly compared visual and multimodal foundation models across several cancer types, demonstrating consistent gains in performance and task adaptability when textual data were incorporated35. However, variability in WSIs quality, staining protocols, scanner devices, and tumor morpho-biological features can compromise model reliability36. This is particularly relevant for rare histologic subtypes, such as micropapillary or apocrine carcinomas which, while underrepresented in large datasets, may possess highly specific morphological features37,38,39. From a deep learning perspective, the morphological distinctiveness might even facilitate tumor recognition by foundation models, potentially reducing the number of training examples required for effective learning40. Nevertheless, the need for ad hoc studies focusing on rare variants remains, especially when the goal is regulatory-grade validation1,41. Beyond model performance, there are systemic barriers to clinical integration that extend to digital infrastructure, data governance, and computational resources42,43. Many pathology departments, particularly in non-academic or resource-limited settings, still operate with fragmented Information Technology (IT) systems and laboratory information systems (LIS) that are ill-suited for AI-based workflows44. These challenges, shared across all AI tools, not just foundation models, are part of the broader effort of digital transition in pathology, which involves standardization of data formats, secure storage, interoperability, and scalable GPU infrastructure45. Recently, it has been emphasized how these challenges can delay or hinder the deployment of AI solutions even in well-resourced centers46,47,48. An overview of recent foundation models and their applications in breast cancer is provided in Table 112,49,50,51,52,53.
Optimized models for breast cancer molecular pathology
In response to the limitations of large-scale foundation models, a new generation of optimized AI models is gaining momentum53,54. These models are intentionally designed to be compact, task-specific, and clinically aligned, offering a pragmatic alternative for AI integration in breast cancer diagnostics35. Rather than attempting to capture the full morphological spectrum of disease, these models are trained to perform well-defined diagnostic or predictive tasks, such as HR, HER2, or Ki-67 status assessment55,56,57, typically in the early-stage setting, where treatment decisions are based on precise immunohistochemical stratification. This task-specific paradigm has been the dominant approach in computational pathology since its inception, well before the advent of foundation models. Although the field has recently embraced large-scale pathology foundation models (PFMs) for their promise of general-purpose adaptability, early experience suggests that their complexity, data requirements, and computational cost may limit their immediate clinical applicability58. Recent work with PFMs has demonstrated strong generalization across multiple tumor types, including rare cancers and out-of-distribution cohorts (AUC ≈ 0.95), but performance gaps remain for certain rare variants, emphasizing that purely generalist solutions may not fully meet clinical needs. These observations underscore the ongoing relevance of task-oriented models, which can be optimized for specific diagnostic or predictive endpoints and deployed efficiently in real-world workflows58. Some models have also been developed to infer actionable genomic alterations, such as germline BRCA mutations, directly from histopathological slides59. Faycal et al. introduced a convolutional neural network (CNN) trained on H&E slides from triple-negative breast cancer cases to predict BRCA mutational status60. Similarly, Bergstrom et al. proposed a deep learning model to predict homologous recombination deficiency (HRD) by integrating histological and genomic features, achieving area under the curve (AUC) values ranging from 0.78 to 0.8761. Rather than directly identifying a unique phenotype, these models estimate the probability of HRD based on morphologic features that co-occur with the alteration in the training set61. As such, they hold promises as screening tools or decision aids to prioritize confirmatory sequencing, particularly in resource-limited settings, but they should not be considered as a replacement for molecular testing.
The benefits of compact models are both technical and clinical. By avoiding the computational burden of large foundation architectures, optimized models offer faster inference times, lower hardware requirements, and greater ease of deployment in real-world pathology workflows62,63. One example is Orpheus, a multimodal deep learning model trained to infer the Oncotype DX Recurrence Score from H&E-stained WSIs64. This tool demonstrated the ability to stratify patients by risk of recurrence, independent of molecular surrogate markers, opening the door to histology-based decision support in settings where molecular assays are unavailable or cost-prohibitive.
These advances extend beyond biomarker prediction. Recently, RlapsRisk BC, a deep learning model developed to assess metastatic relapse risk in early-stage ER-positive/HER2-negative breast cancer, demonstrated that WSIs alone can predict 5-year metastasis-free survival (MFS) with a concordance index (C-index) of 0.81, outperforming traditional clinico-pathological models (C-index 0.76, p < 0.05)65. Notably, combining AI-derived risk with clinical features improved both sensitivity and specificity in patient stratification. Importantly, expert review of model-identified high-impact regions confirmed that the predictions were grounded in recognizable histological features, reinforcing the model’s interpretability and biological plausibility. Together, these examples highlight the clinical promise of optimized AI models for both molecular classification and outcome prediction directly from standard histological slides.
Model distillation and deployment
In breast cancer pathology, the successful adoption of AI depends less on abstract performance metrics and more on its ability to deliver actionable, explainable, and accessible solutions within real-world diagnostic settings66,67,68. Even high-performing foundation models often fail to deliver when they cannot be integrated into routine workflows, explained to clinicians, or accessed by institutions with limited technical infrastructure69,70. Explainability is increasingly recognized as a prerequisite for clinical adoption of AI models, particularly in scenarios where model outputs appear to exceed human perception. Traditional methods such as attention mapping, saliency maps, Grad-CAM, and concept attribution techniques can localize regions or features that most strongly influence model predictions, providing visual cues that support human verification. However, recent evidence has highlighted important limitations. The Explainability Paradox71 showed that different explanation methods may produce inconsistent or even contradictory outputs, and that pathologists vary widely in how they interpret and trust these explanations. Moreover, explainability methods can be sensitive to small perturbations, may highlight non-causal artifacts, and do not necessarily clarify why a given pattern is predictive, raising concerns about stability and fidelity. Beyond explainability, the emerging concept of causability emphasizes that clinicians should be able to interact with AI systems—formulating “what-if” questions, exploring counterfactuals, and investigating how changes in input would alter predictions72. Such interactive and human-in-the-loop approaches may improve understanding, enable error analysis, and foster trust in algorithmic recommendations. This is particularly crucial when AI models appear to outperform human observers, as in the case of the Quantitative Continuous Scoring (QCS) model for TROP2 expression in lung cancer, which provides a reproducible and continuous score that can be cross-validated by experts73. Ensuring that predictions are not only accurate but also interpretable and biologically plausible is essential to support safe deployment and clinician acceptance in real-world workflows. Among the techniques that enable the development of optimized models, distillation stands out for its translational value74,75. Rather than learning from raw data, the distilled model learns from the outputs of the original, inheriting key insights while shedding unnecessary computational weight value74,75. This approach is particularly suited to breast cancer, where molecular features must be interpreted consistently and rapidly across diverse institutional contexts76. Distilled models are easier to interpret, update, and validate, making them well-aligned with regulatory requirements and clinical expectations. Moreover, their simplicity fosters transparency, a prerequisite for clinical trust and broader adoption, especially when AI is used to predict therapeutic biomarkers or to perform risk stratification69,75,77,78. Some examples such as compact models distilled for microsatellite instability (MSI) prediction in colorectal cancer, breast cancer risk estimation directly from H&E slides, and the Quantitative Continuous Scoring (QCS) model for TROP2 expression in lung cancer demonstrate that distillation is more than technical optimization79,80. This approach to TROP2 quantification is also likely to play a role in breast cancer81. The deployment of AI models for diagnostic and predictive biomarker workflows raises several ethical challenges65,82. When algorithms are used to stratify patients for targeted therapies or inclusion in clinical trials, such as with QCS of TROP2 expression, there is a risk that black-box predictions may drive critical clinical decisions without sufficient human interpretability or confirmatory testing. This can amplify biases present in the training data, potentially leading to systematic over- or under-treatment of specific patient subgroups83,84. Ethical deployment therefore requires rigorous external validation, prospective studies, and robust quality control pipelines that allow clinicians to audit model outputs and compare them against visual inspection or orthogonal molecular assays where feasible85. Furthermore, transparent reporting of model development, dataset composition, and performance on diverse populations is essential to ensure equity, reproducibility, and patient safety65,84. These considerations are central to building trustworthy AI systems that complement, rather than replace, expert judgment in pathology.
Bias and fairness considerations
Bias can enter AI-based computational pathology workflows at multiple levels: dataset composition (e.g., over-representation of certain tumor subtypes, demographics, or staining protocols), label quality, model architecture, and evaluation metrics86,87. Importantly, “human-in-the-loop” approaches, while valuable for improving interpretability and trust, can unintentionally amplify bias if the human feedback reflects pre-existing diagnostic conventions or subjective patterns, reinforcing rather than correcting model errors88. Similarly, targeted models optimized for specific biomarkers may perform well in the training domain but fail to generalize to under-represented populations or rare morphologies89. Mitigation strategies include curating diverse and representative training datasets, using bias-aware metrics (e.g., subgroup performance reporting), and performing external validation across multiple institutions84. Regular auditing and monitoring of deployed models are also recommended to detect and correct bias drift over time, ensuring equitable performance for all patient subgroups83,90. Emerging explainability frameworks can further help identify model weaknesses and spurious correlations before deployment91.
Conclusion and future directions: precision over power
AI is entering a new phase in breast cancer pathology, characterized by a shift in focus from technological scale to clinical precision. The limitations of large foundation models, including challenges in integration, interpretability, and consistency across real-world clinical settings, underscore the need for a more pragmatic and clinically oriented approach2,12,69,92. Task-oriented AI models, built through techniques such as model distillation, weak supervision, and modular training, represent a viable and scalable alternative. These optimized systems can support the prediction of key biomarkers, risk stratification, and surrogate molecular signatures, offering a pathway to enhance diagnostic workflows and guide personalized treatment decisions (Fig. 1). Future progress will depend on the quality of the datasets, validation across institutions, and collaboration between computational scientists and pathology teams93,94,95,96. Regulatory clarity and clinical trust are essential to ensure the safe deployment and widespread adoption of AI technologies. Ultimately, by aligning AI development with the specific needs of oncology, the field can progress beyond proof-of-concept stages toward real-world impact, delivering accessible, explainable, and clinically meaningful innovations in breast cancer diagnostics.
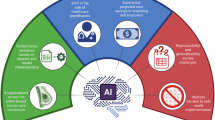
The figure describes the evolution from large foundation models often characterized by unclear “black-box” behavior, high complexity, poor generalizability, to the current focus on more task-oriented, clinically optimized AI systems. The latter systems are designed with a focus on specific diagnostic or predictive objectives, offering clinically interpretable outputs and seamless integration with laboratory information systems (LIS). These models enable more reliable and scalable approaches to the prediction of biomarkers such as HER2-low, Ki-67, PIK3CA, ESR1, and gBRCA. Emerging strategies center on whole-slide image (WSI) analysis, integration with clinical metadata, weak or unsupervised learning, and modular training to enhance real-world performance.
Data availability
No datasets were generated or analyzed during the current study.
References
Soliman, A., Li, Z. & Parwani, A. V. Artificial intelligence’s impact on breast cancer pathology: a literature review. Diagn. Pathol. 19, 38 (2024).
Marra, A. et al. Artificial intelligence entering the pathology arena in oncology: current applications and future perspectives. Ann. Oncol. 36, 712–725 (2025).
Hacking, S. Foundation models in pathology: bridging AI innovation and clinical practice. J. Clin. Pathol. 78, 433–435 (2025).
Chen, W. et al. A visual-omics foundation model to bridge histopathology with spatial transcriptomics. Nat. Methods 22, 1568–1582 (2025).
Ochi, M., Komura, D. & Ishikawa, S. Pathology foundation models. JMA J. 8, 121–130 (2025).
Campanella, G. et al. Real-world deployment of a fine-tuned pathology foundation model for lung cancer biomarker detection. Nat. Med. https://doi.org/10.1038/s41591-025-03780-x (2025).
Fusco, N. et al. HER2 in gastric cancer: a digital image analysis in pre-neoplastic, primary and metastatic lesions. Mod. Pathol. 26, 816–824 (2013).
Gevaert, O. et al. Evaluating vision and pathology foundation models for computational pathology: a comprehensive benchmark study. Res. Sq. https://doi.org/10.21203/rs.3.rs-6823810/v1 (2025).
Albahri, M. et al. A new approach combining a whole-slide foundation model and gradient boosting for predicting BRAF mutation status in dermatopathology. Comput. Struct. Biotechnol. J. 27, 2503–2514 (2025).
Jang, W. et al. Molecular classification of breast cancer using weakly supervised learning. Cancer Res. Treat. 57, 116–125 (2025).
Campanella, G. et al. Computational pathology at health system scale – self-supervised foundation models from three billion images. Preprint at https://arxiv.org/abs/2310.07033 (2023).
Xu, H., et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Jahanifar, M. et al. Domain Generalization in Computational Pathology: Surveyand Guidelines. ACM Comput. Surv. 57, 37 (2025).
Kerle, I. A. et al. Translational and clinical comparison of whole genome and transcriptome to panel sequencing in precision oncology. NPJ Precis. Oncol. 9, 9 (2025).
Venetis, K. et al. ESR1 testing on FFPE samples from metastatic lesions in HR + /HER2- breast cancer after progression on CDK4/6 inhibitor therapy. Breast Cancer Res. 27, 79 (2025).
Chaki, M. et al. Retrospective comparison between breast cancer tissue- and blood-based next-generation sequencing results in detection of PIK3CA, AKT1, and PTEN alterations. Breast Cancer Res. 27, 122 (2025).
Shah, N. M. & Meric-Bernstam, F. The present and future of precision oncology and tumor-agnostic therapeutic approaches. Oncologist 30, (2025).
Gazola, A. A., Lautert-Dutra, W., Archangelo, L. F., Reis, R. B. D. & Squire, J. A. Precision oncology platforms: practical strategies for genomic database utilization in cancer treatment. Mol. Cytogenet. 17, 28 (2024).
Song, A. H. et al. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng. 1, 930–949 (2023).
Brancato, V. et al. Standardizing digital biobanks: integrating imaging, genomic, and clinical data for precision medicine. J. Transl. Med. 22, 136 (2024).
Zarella, M. D. et al. Artificial intelligence and digital pathology: clinical promise and deployment considerations. J. Med. Imaging 10, 051802 (2023).
Frascarelli, C. et al. Revolutionizing cancer research: the impact of artificial intelligence in digital biobanking. J. Pers. Med. 13, https://doi.org/10.3390/jpm13091390 (2023).
Bonizzi, G., Zattoni, L. & Fusco, N. Biobanking in the digital pathology era. Oncol. Res. 29, 229–233 (2021).
Evans, H. et al. Standardized clinical annotation of digital histopathology slides at the point of diagnosis. Mod. Pathol. 36, 100297 (2023).
Friends of Cancer Research. Considerations for Developing Reference Data Sets for Digital Pathology Biomarkers (Friends of Cancer Research, 2025).
McGenity, C. et al. Artificial intelligence in digital pathology: a systematic review and meta-analysis of diagnostic test accuracy. npj Digital Med. 7, 114 (2024).
Lan, Y. C. et al. Ecologically sustainable benchmarking of AI models for histopathology. npj Digital Med. 7, 378 (2024).
Matias-Guiu, X. et al. Implementing digital pathology: qualitative and financial insights from eight leading European laboratories. Virchows Arch. https://doi.org/10.1007/s00428-025-04064-y (2025).
Datwani, S., Khan, H., Niazi, M. K. K., Parwani, A. V. & Li, Z. Artificial intelligence in breast pathology: Overview and recent updates. Hum. Pathol. 105819, https://doi.org/10.1016/j.humpath.2025.105819 (2025).
Al-Zoghby, A. M., Ismail Ebada, A., Saleh, A. S., Abdelhay, M. & Awad, W. A. A Comprehensive Review of Multimodal Deep Learning for Enhanced Medical Diagnostics. Comput. Mater. Contin. 84, 4155–4193 (2025)..
Jarkman, S. et al. Generalization of deep learning in digital pathology: experience in breast cancer metastasis detection. Cancers 14, https://doi.org/10.3390/cancers14215424 (2022).
Ferber, D., et al. In-context learning enables multimodal large language models to classify cancer pathology images. Nat. Commun. 15, 10104 (2024).
Munzone, E. et al. Development and validation of a natural language processing algorithm for extracting clinical and pathological features of breast cancer from pathology reports. JCO Clin. Cancer Inf. 8, e2400034 (2024).
Zhang, S. et al. A Multimodal biomedical foundation model trained from fifteen million image–text pairs. NEJM AI 2, https://doi.org/10.1056/AIoa2400640 (2025).
Chen, J., et al. A deep learning-based multimodal medical imaging model for breast cancer screening. Sci. Rep. 15, 14696 (2025).
Tellez, D., et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 58, 101544 (2019).
Marchio, C., Pietribiasi, F., Castiglione, R., Fusco, N. & Sapino, A. Giants in a microcosm”: multinucleated giant cells populating an invasive micropapillary carcinoma of the breast. Int. J. Surg. Pathol. 23, 654–655 (2015).
Sciarra, A., et al. Columnar cell lesion and apocrine hyperplasia of the breast: is there a common origin? The role of prolactin-induced protein. Appl Immunohistochem. Mol. Morphol. 27, 508–514 (2019).
Venetis, K. et al. The molecular landscape of breast mucoepidermoid carcinoma. Cancer Med. (2023).
L’Imperio, V. et al. Machine learning streamlines the morphometric characterization and multiclass segmentation of nuclei in different follicular thyroid lesions: everything in a NUTSHELL. Mod. Pathol. 37, 100608 (2024).
Stathonikos, N., Nguyen, T. Q., Spoto, C. P., Verdaasdonk, M. A. M. & van Diest, P. J. Being fully digital: perspective of a Dutch academic pathology laboratory. Histopathology 75, 621–635 (2019).
Sanchini, V. et al. A comprehensive ethics and data governance framework for data-intensive health research: Lessons from an Italian cancer research institute. Account Res. 1–18, https://doi.org/10.1080/08989621.2023.2248884 (2023).
Nansumba, H., Ssewanyana, I., Tai, M. & Wassenaar, D. Role of a regulatory and governance framework in human biological materials and data sharing in National Biobanks: case studies from Biobank Integrating Platform, Taiwan and the National Biorepository, Uganda. Wellcome Open Res. 4, 171 (2019).
Zeng, Z., Yi, Z. & Xu, B. The biological and technical challenges facing utilizing circulating tumor DNA in non-metastatic breast cancer patients. Cancer Lett. 616, 217574 (2025).
Buchta, C. et al. Behind the scenes of EQA - characteristics, capabilities, benefits and assets of external quality assessment (EQA). Clin. Chem. Lab. Med. https://doi.org/10.1515/cclm-2024-1289 (2025).
Lu, M. Y., et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021).
Sandbank, J., et al. Validation and real-world clinical application of an artificial intelligence algorithm for breast cancer detection in biopsies. NPJ Breast Cancer 8, 129 (2022).
Dunenova, G. et al. The performance and clinical applicability of HER2 digital image analysis in breast cancer: a systematic review. Cancers 16, https://doi.org/10.3390/cancers16152761 (2024).
Lu, M. Y., et al. A visual-language foundation model for computational pathology. Nat. Med. 30, 863–874 (2024).
Wu, X., et al. Multimodal recurrence risk prediction model for HR+/HER2- early breast cancer following adjuvant chemo-endocrine therapy: integrating pathology image and clinicalpathological features. Breast Cancer Res. 27, 27 (2025).
Campanella, G., et al. A clinical benchmark of public self-supervised pathology foundation models. Nat. Commun. 16, 3640 (2025).
Filiot, A., Jacob, P., Kain, A. M. & Saillard, C. Phikon-v2, A large and public feature extractor for biomarker prediction. ArXiv, abs/2409.09173 (2024).
Wang, X., et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 81, 102559 (2022).
Chen, R. J., et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 30, 850–862 (2024).
Zhang, M., et al. Developing a weakly supervised deep learning framework for breast cancer diagnosis with HR status based on mammography images. Comput. Struct. Biotechnol. J. 22, 17–26 (2023).
Farahmand, S., et al. Deep learning trained on hematoxylin and eosin tumor region of Interest predicts HER2 status and trastuzumab treatment response in HER2+ breast cancer. Mod. Pathol. 35, 44–51 (2022).
Matsumoto, H., et al. Ki-67 evaluation using deep-learning model-assisted digital image analysis in breast cancer. Histopathology 86, 460–471 (2025).
Vorontsov, E., et al. A foundation model for clinical-grade computational pathology and rare cancers detection. Nat. Med. 30, 2924–2935 (2024).
Li, Y., et al. An interpretable deep learning model for detecting BRCA pathogenic variants of breast cancer from hematoxylin and eosin-stained pathological images. PeerJ 12, e18098 (2024).
Faycal, T., Gaceb, D., Belkadi, C. & Loubar, B. A Self-Supervised Learning Approach for Detecting BRCA Mutations in Breast Cancer Histopathological Images. In Proceedings 7th International Conference on Informatics & Data-Driven Medicine (IDDM 2024); Vol. 3892, 183−195 (CEUR Workshop Proceedings, 2025).
Bergstrom, E. N., et al. Deep learning artificial intelligence predicts homologous recombination deficiency and platinum response from histologic slides. J. Clin. Oncol. 42, 3550–3560 (2024).
Baxi, V., Edwards, R., Montalto, M. & Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 35, 23–32 (2022).
Janowczyk, A. & Madabhushi, A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inf. 7, 29 (2016).
Boehm, K. M., et al. Multimodal histopathologic models stratify hormone receptor-positive early breast cancer. Nat. Commun. 16, 2106 (2025).
Chauhan, C. & Gullapalli, R. R. Ethics of AI in pathology: current paradigms and emerging issues. Am. J. Pathol. 191, 1673–1683 (2021).
Pierce, R. L., Van Biesen, W., Van Cauwenberge, D., Decruyenaere, J. & Sterckx, S. Explainability in medicine in an era of AI-based clinical decision support systems. Front. Genet. 13, 903600 (2022).
Tiwari, A., Mishra, S. & Kuo, T. R. Current AI technologies in cancer diagnostics and treatment. Mol. Cancer 24, 159 (2025).
Echle, A., et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer 124, 686–696 (2021).
Stenzinger, A., et al. Artificial intelligence and pathology: from principles to practice and future applications in histomorphology and molecular profiling. Semin. Cancer Biol. 84, 129–143 (2022).
Kiani, A., et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 3, 23 (2020).
Evans, T. et al. The explainability paradox: challenges for xAI in digital pathology. Fut. Gener. Comput. Syst. 133, https://doi.org/10.1016/j.future.2022.03.009 (2022).
Plass, M. et al. Explainability and causability in digital pathology. J. Pathol. Clin. Res. 9, 251–260 (2023).
Garassino, M. C., et al. PL02.11 normalized membrane ratio of TROP2 by quantitative continuous scoring is predictive of clinical outcomes in TROPION-Lung 01. J. Thorac. Oncol. 19, S2–S3 (2024).
Alkhulaifi, A., Alsahli, F. & Ahmad, I. Knowledge distillation in deep learning and its applications. PeerJ Comput. Sci. 7, e474 (2021).
Hu, Y., et al. Analysis of genomic and proteomic data using advanced literature mining. J. Proteome Res. 2, 405–412 (2003).
Guo, W. et al. Self-knowledge distillation for prediction of breast cancer molecular subtypes based on digital breast tomosynthesis. Med. Biol. Eng. Comput. https://doi.org/10.1007/s11517-025-03383-1 (2025).
Teng, Q., Liu, Z., Song, Y., Han, K. & Lu, Y. A survey on the interpretability of deep learning in medical diagnosis. Multimed. Syst. 28, 2335–2355 (2022).
Pesapane, F., et al. Advances in breast cancer risk modeling: integrating clinics, imaging, pathology and artificial intelligence for personalized risk assessment. Fut. Oncol. 19, 2547–2564 (2023).
Cao, R., et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 10, 11080–11091 (2020).
Chaudhury, S., Shelke, N., Sau, K., Prasanalakshmi, B. & Shabaz, M. A novel approach to classifying breast cancer histopathology biopsy images using bilateral knowledge distillation and label smoothing regularization. Comput. Math. Methods Med. 2021, 4019358 (2021).
Cursano, G., et al. Trop-2 as an actionable biomarker in breast cancer. Curr. Genomics 24, 129–131 (2023).
McKay, F., et al. The ethical challenges of artificial intelligence-driven digital pathology. J. Pathol. Clin. Res. 8, 209–216 (2022).
Hanna, M. G., et al. Ethical and bias considerations in artificial intelligence/machine learning. Mod. Pathol. 38, 100686 (2025).
Hasanzadeh, F., et al. Bias recognition and mitigation strategies in artificial intelligence healthcare applications. npj Digital Med. 8, 154 (2025).
Matthews, G. A., McGenity, C., Bansal, D. & Treanor, D. Public evidence on AI products for digital pathology. npj Digital Med. 7, 300 (2024).
Koçak, B., et al. Bias in artificial intelligence for medical imaging: fundamentals, detection, avoidance, mitigation, challenges, ethics, and prospects. Diagn. Inter. Radio. 31, 75–88 (2025).
Cross, J. L., Choma, M. A. & Onofrey, J. A. Bias in medical AI: implications for clinical decision-making. PLOS Digit Health 3, e0000651 (2024).
Rosbach, E., Ganz, J., Ammeling, J., Riener, A. & Aubreville, M. Automation Bias in AI-assisted Medical Decision-making under Time Pressure in Computational Pathology. In Bildverarbeitung für die Medizin 2025. BVM 2025. Informatik aktuell. (ed. Palm, C. et al.) (Springer Vieweg, Wiesbaden, 2025).
Liu, M., et al. A scoping review and evidence gap analysis of clinical AI fairness. NPJ Digit Med. 8, 360 (2025).
Jackson, B. R. et al. The ethics of artificial intelligence in pathology and laboratory medicine: principles and practice. Acad. Pathol. 8, https://doi.org/10.1177/2374289521990784 (2021).
Kaczmarzyk, J. R., Saltz, J. H. & Koo, P. K. Explainable AI for computational pathology identifies model limitations and tissue biomarkers. Preprint at https://arxiv.org/abs/2409.03080 (2024).
Klauschen, F., et al. Toward explainable artificial intelligence for precision pathology. Annu. Rev. Pathol. 19, 541–570 (2024).
Frascarelli, C., et al. Deep learning algorithm on H&E whole slide images to characterize TP53 alterations frequency and spatial distribution in breast cancer. Comput. Struct. Biotechnol. J. 23, 4252–4259 (2024).
Ehteshami Bejnordi, B., et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017).
Caldonazzi, N. et al. Value of artificial intelligence in evaluating lymph node metastases. Cancers 15, https://doi.org/10.3390/cancers15092491 (2023).
Sajjadi, E. et al. Computational pathology to improve biomarker testing in breast cancer: how close are we? Eur. J. Cancer Prev. https://doi.org/10.1097/cej.0000000000000804 (2023).
Acknowledgements
Konstantinos Venetis was supported by the Fondazione Umberto Veronesi; The final proofreading of grammar and syntax for the manuscript was conducted using ChatGPT 5 and Grammarly v.6.8.263. All content development, interpretation, and scientific conclusions are solely the responsibility of the authors. This work was partially supported by the Italian Ministry of Health through Ricerca Corrente 5 × 1000 funds; the Italian Ministry of Innovations via the Sustainable Growth Fund – Innovation Agreements under the Ministerial Decree of December 31, 2021, and the Director's Decree of November 14, 2022 (2nd Call), Project No.: F/350104/01–02/X60; and the Italian Ministry of University and Research (MUR) 2023 through the “Future Artificial Intelligence Research – FAIR” program, PE0000013, CUP D53C22002380006, within the National Recovery and Resilience Plan (PNRR), Mission 4, Component 2, Investment 1.3 – funded by the European Union – NextGenerationEU. Project: “AIDH – Adaptive AI Methods for Digital Health.
Author information
Authors and Affiliations
Contributions
Study conception and design, N.F.; methodology (search and selection criteria for the references), K.V. and C.F.; writing - original draft preparation, N.F.; writing - review and editing, K.V., C.F., A.C., E.M., F.P., M.D.E., S.K., M.N.; revision, A.M., G.C.; figure draft, C.F., A.C., E.M., F.P., M.D.E., S.K., M.N.; supervision, K.V., N.F.; project administration, E.G.-R., N.F.
Corresponding author
Ethics declarations
Competing interests
K.V. Has received honoraria for speaker bureau from Merck Sharp & Dohme (MSD), Roche, and AstraZeneca; A.M. has received support from Menarini Group and served on the Speakers’ Bureau for Roche and AstraZeneca. G.C. has received honoraria for speaker engagements from Roche, Seattle Genetics, Novartis, Lilly, Pfizer, Foundation Medicine, NanoString, Samsung, Celltrion, BMS, and MSD; honoraria for consultancy from Roche, Seattle Genetics, and NanoString; honoraria for participation in advisory boards from Roche, Lilly, Pfizer, Foundation Medicine, Samsung, Celltrion, and Mylan; honoraria for writing engagements from Novartis and BMS; and honoraria for participation in the Ellipsis Scientific Affairs Group. He has also received institutional research funding for conducting phase I and II clinical trials from Pfizer, Roche, Novartis, Sanofi, Celgene, Servier, Orion, AstraZeneca, Seattle Genetics, AbbVie, Tesaro, BMS, Merck Serono, Merck Sharp & Dohme, Janssen-Cilag, Philogen, Bayer, Medivation, and Medimmune. E.G-R. has received advisory fees, honoraria, travel accommodations/expenses, grants, and/or non-financial support from AstraZeneca, Exact Sciences, GSK, Illumina, MSD, Novartis, Roche, and Thermo Fisher Scientific; N.F. has received honoraria for consulting, advisory role, speaker bureau, travel, and/or research grants from Abbvie, Alira Health, AstraZeneca, Daiichi Sankyo, Epredia, Exact Sciences, Gilead, GSK, Leica Biosystems, Lilly, Menarini Group, Merck, MSD, Novartis, Pfizer, Roche, Sakura, Sysmex, ThermoFisher, Veracyte. These companies had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and/or in the decision to publish the results. All other authors declare no potential conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Frascarelli, C., Venetis, K., Marra, A. et al. Computational pathology in breast cancer: optimizing molecular prediction through task-oriented AI models. npj Breast Cancer 11, 141 (2025). https://doi.org/10.1038/s41523-025-00857-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41523-025-00857-1