Abstract
Hormone receptor-positive (HR + )/HER2-negative (HER2 − ) early breast cancers (BCs) are typically considered immunologically cold. However, combining immune checkpoint inhibitors (ICIs) with chemotherapy has shown to improve pathological complete response (pCR) in high-risk patients. Understanding the relationship between immune activation and tumor biology may help identify HR + /HER2 − BC patients most likely to benefit from such combinations. Baseline gene expression data from two neoadjuvant trials (GIADA and LETLOB) including HR + /HER2 − BC patients were analyzed. PAM50 intrinsic subtyping and relevant immune-related gene signatures were calculated. Tumor-infiltrating lymphocytes (TILs) were assessed on baseline samples. Among 109 tumors, PAM50 classified 44% as Luminal-B (LumB), 33% Luminal-A (LumA), 18% Basal-like, and 5% HER2-enriched. TIL levels (available for N = 101) were generally low (median 2; range 0–100), with higher levels in Basal-like BCs (p = 0.008). Basal-like BCs exhibited significantly higher levels of immune-related signatures (CD8 T-cells, Cytotoxic cells, IFN-γ, response to ICI + CT in GeparNuevo) and of PD-1, PD-L1, and PD-L2 genes. No differences were found between LumA and LumB subtypes. TILs and immune signatures showed a significant weak-to-moderate positive correlation with the basal-like signature. HR + /HER2- BCs that display both features of biological aggressiveness and enhanced immunogenicity may represent ideal candidates for the combination of ICI and chemotherapy.
Similar content being viewed by others
Introduction
Hormone receptor-positive (HR + )/HER2- breast cancer (BC) is the most common BC subtype, accounting for approximately 70% of BC diagnoses worldwide. HR + /HER2- BC is an extremely heterogeneous disease, as has been progressively elucidated by a deeper understanding of its biological complexity over the past decades. This knowledge has directly influenced current clinical practice, as the introduction of genomic assays and the use of endocrine “window-of-opportunity” studies—assessing Ki67 response to short-term endocrine therapy—have allowed a more precise assessment of tumor biology. This has led to an increased personalization of treatment management of early HR + /HER2- BC, with escalation and de-escalation strategies available based on individual patient risk and tumor features1,2.
From a biological point of view, gene expression profiling can be used to explore the biological heterogeneity of BC with the identification of four ‘intrinsic subtypes’, namely luminal A (LumA), luminal B (LumB), HER2-enriched (HER2-E), basal-like (BL) BC, and a normal-like group3. Several studies have demonstrated that, although in different proportions, all PAM50 intrinsic subtypes can be detected within each immunohistochemistry-based subgroup, and that around 10% of early-stage HR + /HER2- early BCs are classified as non-luminal (e.g., HER2-E and BL) by gene expression. In addition, these subtypes significantly differ in terms of prognosis and treatment response. It has been consistently reported that, within HR + /HER2- early BC, tumors classified as basal-like by gene expression present a worse prognosis3,4, and are associated with higher sensitivity to chemotherapy and lower sensitivity to endocrine treatment4,5,6,7,8.
While the addition of immunotherapy to neoadjuvant chemotherapy has become standard practice for high-risk triple negative BC (TNBC), HR + /HER2- BCs have traditionally been considered immunologically ‘cold’ tumors2,9. Nevertheless, two phase III trials have recently reported an increase in pathologic complete response (pCR) rates with the addition of immune checkpoint inhibitor to standard chemotherapy in patients with high-risk HR + /HER2- BC, suggesting a potential role for immunotherapy in at least a subgroup of early/locally advanced HR + /HER2- BC10,11. A biomarker analysis of the CheckMate 7FL trial, assessing the addition of nivolumab to standard chemotherapy in patients with high-risk HR + /HER2- BC, reported that both stromal tumor infiltrating lymphocytes (TILs) levels (>1% or ≥5%) and PD-L1 levels (defined as IC ≥ 1% or CPS ≥ 3) were independently associated with nivolumab efficacy in this patient population, highlighting the potential relevance of immune-based biomarkers in this setting11.
Therefore, both intrinsic tumor biology and immune microenvironment characteristics might play a role in modulating response to specific treatments in HR + /HER2- BC. Indeed, previous data from a phase II trial identified both basal-like molecular subtype and TILs as biomarkers independently associated with the benefit of neoadjuvant immunotherapy and chemotherapy in HR + /HER2- BC12. Moreover, within HR + /HER2- subtype, TILs may present a different prognostic role according to tumor biological features. While there is sparse available evidence suggesting a marginal clinical value of TILs quantification in terms of prognostic stratification in unselected HR + /HER2- BC patients, more consistent data is available regarding a positive prognostic role of TILs in patients whose HR + /HER2- tumors exhibit features of higher biological aggressiveness13,14. Thus, there is an interest in assessing the interaction between immune features and tumor biology in HR + /HER2- BC to identify subsets of patients who could be ideal candidates for the combination of chemotherapy and immune checkpoint inhibitors.
In this study, we aimed to assess, in early HR + HER2- BC, the interplay between tumor biology, as assessed by gene expression, and immune microenvironment features, assessed by TILs evaluation and immune genomic signatures (i.e., CD8 T-cells, cytotoxic cells, IFN-γ–activation) previously reported to be correlated with immunotherapy benefit across several cancer types15. To this end, we performed a joint analysis of translational data from two multicentric neoadjuvant phase II trials (the GIADA trial and the LETLOB trial). Together, these trials provide a quite comprehensive representation of patients with HR + /HER2– BC who are eligible for neoadjuvant treatment. Specifically, the GIADA trial enrolled premenopausal patients diagnosed with HR + /HER2- BC classified as luminal-B like by immunohistochemistry who were treated with chemotherapy, endocrine therapy, and nivolumab, while the LETLOB trial included postmenopausal patients with HR + /HER2- BC and eligible for neoadjuvant treatment who received neoadjuvant letrozole with or without lapatinib. Collectively, these cohorts reflect the heterogeneity of the HR + /HER2– population encountered in clinical practice, with the GIADA trial offering a particularly robust representation of luminal B–like disease.
Results
Clinical patient characteristics
Overall, 109 patients were included in the present study (43 from the GIADA trial and 66 from the LETLOB trial).
Main clinicopathological characteristics of study population are reported in Table 1. It is noteworthy that ER expression was high in this patient cohort (median value 95%, range 25–100%) and the majority of patients also presented PR-positive tumors (median value 73%, range 0–100%).
The majority of patients presented tumors with histological grade 2 or grade 3, with fewer than 1% of cases classified as grade 1. More than half of the patients included presented a stage IIA disease (53.2%), while the remaining patients presented either a stage IIB (35.8%) or stage IIIA (11%) tumor.
PAM50 subtyping: overall and association with clinicopathological features
Gene expression data were available for all patients included in the study.
According to PAM50 intrinsic subtyping, most patients presented a Luminal-like BC (Luminal A in 33% of cases, Luminal B in 44%), while 18% presented a basal-like subtype and 5% a HER2-enriched subtype.
Basal-like, HER2-enriched and LumB subtypes were more frequently observed in histological grade 3 tumors (23.5%, 7.8%, and 49.0%, respectively) than in histological grade 2 tumors (12.8%, 2.1%, and 36.1% respectively), while LumA subtypes were less frequently observed in histological grade 3 tumors (19.6%) than in histological grade 2 tumors (48.9%) (overall p = 0.016). In addition, a significant difference in Ki67 expression levels was observed according to PAM50 intrinsic subtype, with Basal-like tumors showing the highest values and LumA tumors showing the lowest values of Ki67 (p < 0.001, Supplementary Table 1), while no significant difference in ER expression levels (p = 0.439) and PgR expression levels was observed (p = 0.089) was observed.
No significant difference in PAM50 subtype distribution was observed according to tumor stage (p = 0.478) and menopausal status (p = 0.082) (Supplementary Table 1).
TIL evaluation and association with clinicopathological features
TIL data were available for 101 patients included in the study.
As expected for HR + /HER2- BC, TIL levels were generally low (median value 2, with 83.2% of cases with evaluable TILs classified as ≤10%); however, a significant variability was observed with 7.3% of cases (N = 8) classified as TILs ≥30%.
When the distribution of TIL levels across different clinicopathological subgroups was assessed, nostrong association between TIL levels and clinicopathological features was observed. Indeed, no significant difference in TIL levels was observed according to histological grade (p = 0.703), tumor stage (p = 0.716), and menopausal status (p = 0.355) (Supplementary Fig. 1), Additionally, no significant correlation was observed between TIL levels and ER levels (Spearman Rho = −0.159, p = 0.112), and only a weak negative association was observed between TIL levels and PgR levels (Spearman’s Rho = −0.245, p = 0.014) and a weak positive association between TIL levels and Ki67 levels (Spearman’s Rho = 0.202, p = 0.044) was observed.
TIL levels according to PAM50 subtyping
To further explore the potential interplay between tumor biology and TIL levels, we assessed their association with PAM50 subtypes.
A significant association between TIL levels and PAM50 intrinsic subtypes was observed, with basal-like HR + /HER2- BCs showing significantly higher TIL levels as compared to other subtypes (overall p = 0.008, Fig. 1). In particular, a significant difference in TIL levels was observed between Basal-like tumors and LumA tumors (p = 0.019) and a trend towards higher TIL levels for Basal-like tumors was observed when compared to LumB tumors (p = 0.055), while no difference was observed between LumA and LumB tumors (p = 0.532). Due to the very limited number of cases classified as HER2-enriched (N = 5), no comparison was made with this subgroup.
To assess the association between more subtle basal-like tumor biology features and immune activation as assessed by TIL levels, we also assessed the basal-like PAM50 signature as a continuous variable. Consistently with the previous observation, a weak positive correlation was observed between continuous TIL levels and the basal-like PAM50 signature assessed as a continuous variable (Spearman’s Rho=0.372, p < 0.001; Fig. 2A), while a weak negative correlation was observed between continuous TILs and the LumA PAM50 signature when assessed as a continuous variable (Spearman’s Rho = −0.372, p < 0.001; Supplementary Fig. 2).
Immune signature expression according to PAM50 subtyping
To better evaluate the potential interplay between tumor biology and immune regulation, previously published immune signatures associated with CD8 T-cells and Cytotoxic cells, IFN-gamma and an immune gene signature previously response to ICI in the GeparNuevo trial were computed (109 patients with available information).
A significant association between PAM50 intrinsic subtypes and each of the four immune signatures was observed, with basal-like HR + /HER2- BCs showing significantly higher levels of all the 4 immune signatures as compared to the other subtypes (p < 0.001, Fig. 3).
Additionally, the basal-like PAM50 signature (as a continuous variable) was the only PAM50 gene signature showing a weak-to-moderate positive correlation with each of the four immune signatures evaluated (p < 0.001, Fig. 2B–E; detailed data for all PAM50 signatures are reported in Supplementary Table 2).
Expression of significant immune genes according to PAM50 subtyping
Only for patients included in the GIADA trial (43 patients), the baseline expression of genes codifying for PD-1 (PDCD1), PD-L1 (CD274) and PD-L2 (PDCD1-lg2) was also available. Again, significantly higher mRNA levels of all these three genes were observed in basal-like HR + /HER2- BCs as compared to the other subtypes (p = 0.002 for PDCD1, and p < 0.001 for CD274 and PDCD1-lg2; Fig. 4). Consistently, a moderate-to-strong positive correlation was observed between the mRNA levels of these three genes and the basal-like PAM50 signature assessed as a continuous variable, while a weak to moderate negative correlation was observed between the mRNA levels of these three genes and the LumA and the LumB PAM50 signatures assessed as continuous variables (detailed data for all PAM50 signatures are reported in Supplementary Table 3).
Expression of PD-1 and PD-L1 assessed by immunofluorescence according to PAM50 subtyping
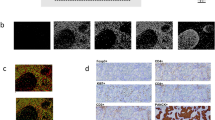
Only for patients included in the GIADA trial (43 patients), baseline expression of PD-1 and PD-L1 expression on T lymphocytes, macrophages, and tumor cells was assessed on FFPE tumor biopsies using a mIF approach. Consistent with previous results, a significantly higher density of CD3 + PD-1+ cells (p = 0.010) and a significantly higher density of PD-L1+ cells (p = 0.043) in the tumor area were detected in basal-like HR + /HER2– BC as compared to other intrinsic subtypes (Supplementary Figs. 3–4). As expected, this was mainly related to a significantly higher density of PD-L1+ macrophages (CD68 + ) (p = 0.043) in the tumor area of basal-like HR + /HER2– BC as compared to other intrinsic subtypes, while we did not observe significant differences in the density of PD-L1+ tumor cells (pan-cytokeratin + ) in the tumor area according to intrinsic subtype (p = 0.222) (Supplementary Figs. 5–6). Coherently, the density of CD3 + PD-1+ cells, PD-L1+ cells, and more specifically PD-L1 + CD68+ cells in the tumor area showed a moderate positive correlation with the basal-like PAM50 signature, when assessed as a continuous variable. Conversely, a weak-to-moderate negative correlation were observed between these immune markers and the LumA PAM50 signature assessed as a continuous variable (detailed data for all PAM50 signatures are provided in Supplementary Table 4).
Discussion
HR + /HER2- has traditionally been described as an immunologically “cold” tumor, characterized by low levels of TILs and reduced immune activation16. However, emerging evidence suggests that immunotherapy, in combination with neoadjuvant chemotherapy, may provide significant benefits for at least a subgroup of patients within HR + /HER2- BC. Indeed, in the CheckMate 7FL trial, the addition of nivolumab to neoadjuvant chemotherapy improved pCR rates by 10.5% as compared to chemotherapy alone11. Similarly, the KEYNOTE 756 trial reported an 8.5% increase in pCR rates with the addition of pembrolizumab to neoadjuvant chemotherapy10. These findings highlight the potential efficacy of chemoimmunotherapy in HR + /HER2- BC; however, given the potential long-term toxicities associated with this approach, also underscore the critical need for biomarkers to better identify which patients are more likely to benefit.
In this context, some biomarkers have recently shown promise. While immune-based biomarkers have been assessed in the CheckMate 7FL trial, showing that both stromal tumor infiltrating lymphocytes (TILs) levels (>1% or ≥5%) and PD-L1 levels (defined as IC ≥ 1% or CPS ≥ 3) were independently associated with nivolumab efficacy, other studies have also taken into account tumor biology. In particular, in the GIADA trial, in which premenopausal patients with Luminal B-like (by IHC) BCs were treated with chemotherapy, immunotherapy and endocrine treatment identified the PAM50 Basal-like intrinsic subtype and high levels of TILs as independently associated with higher pCR rates. A composite score combining these features demonstrated excellent predictive accuracy, with an area under the curve (AUC) of 0.9512. Moreover, in the I-SPY2 trial, HR + /HER2- BC patients with MammaPrint-High2 tumors or BluePrint Basal-type tumors showed marked increased pCR rates with the addition of pembrolizumab to a taxane-based regimen17, and were also associated with significantly worse long-term outcomes in patients not achieving a pCR18. In addition, transcriptomic analyses from this study identified a 53-gene signature (mainly involving immune genes), termed ImPrint, which was strongly associated with increase in pCR rates with the addition of pembrolizumab to neoadjuvant chemotherapy both in HR + /HER2- BC and in triple-negative BC, but did not show a significant impact on long-term outcomes in patients not achieving a pCR19. Interestingly, only a partial overlap between the MammaPrint-High2, BluePrint Basal-type, and ImPrint+ signatures was observed in this study17.
Our study adds to the existing evidence in this context by further exploring the association between TILs, immune signatures, and tumor biology features as assessed by PAM50 intrinsic subtyping in HR + /HER2- early BC. Indeed, in our cohort of HR + /HER2- BCs, basal-like tumors showed higher TILs level and higher levels of all four immune signatures tested in this study20,21. This observation supports the hypothesis that some biological features associated with aggressive clinical behavior in HR + HER2- BC might also influence the immunogenicity of the tumor and shape the composition of the immune microenvironment.
Despite the potential clinical significance, a comprehensive understanding of the interplay between tumor biology and immune features in HR + /HER2- BC remains elusive. Most efforts to date have been primarily focused on either tumor-intrinsic features or immune features, with limited data integrating these distinct biological domains. In our study, only a weak positive correlation being observed between continuous TILs and the basal-like PAM50 signature and a weak to moderate positive correlation with the four immune signatures was identified. This indicates that PAM50 subtyping and immunological markers, such as TILs and immune signatures, capture only partially overlapping biological information. Considering the well-established association between the PAM50 Basal-like subtype and worse prognosis in HR + /HER2- BC, and taking into account its higher sensitivity to chemotherapy, patients whose tumors exhibit both aggressive biological features and enhanced immunogenicity may represent ideal candidates for neoadjuvant chemoimmunotherapy.
Furthermore, while the previously cited phase III trials testing the use of neoadjuvant chemoimmunotherapy in HR + /HER2- BC mainly included patients with grade 3 tumors, our analysis also encompassed a significant number of patients with grade 2 tumors. Despite this, no significant association was observed between histological grade and TIL levels.
This study has several limitations. The analyses were based on non-randomized trials with small sample sizes and different inclusion criteria. Indeed, the limited sample size of the present study, and particularly the limited number of patients exhibiting specific biomarkers (e.g., basal-like tumors) may limit the robustness and generalizability of our findings. Therefore, our results should be considered hypothesis-generating. In addition, the correlation between basal-like subtype and PD-1, PD-L1 and PD-L2 mRNA levels could be assessed only in samples derived from the GIADA study and should thus be interpreted with additional caution, given the limited number of patients and the specific inclusion criteria of the study, further limiting generalizability. Tissue samples were not consistently evaluable across all patients, and gene expression analyses were performed using different technologies, potentially affecting comparability.
Overall, while HR + /HER2- BC remains a challenging subtype for immunotherapy, emerging evidence supports the potential of immune checkpoint inhibitors in combination with chemotherapy for selected patients. In this context, our results emphasize the need for the integration of biomarkers that integrate both tumor and immune features in clinical trial design to facilitate personalized treatment strategies, maximizing therapeutic efficacy while minimizing unnecessary toxicities. Future research should focus on validating these biomarkers using standardized methodologies in larger, randomized trials.
Methods
GIADA neoadjuvant trial cohort
The GIADA trial (NCT04659551, EUDRACT 2016-004665-10) is an investigator-driven, multicentric, phase II, single-arm trial, which included premenopausal women with newly diagnosed, previously untreated, stage II-IIIA HR + /HER2- BC classified as LumB-like by immunohistochemistry (Ki67 ≥ 20% and/or histologic grade 3) and who were candidate for neoadjuvant chemotherapy based on local multidisciplinary evaluation.
Patients enrolled in the GIADA trial received three 21-day cycles of intravenous EC (epirubicin 90 mg/m2 and cyclophosphamide 600 mg/m2), followed by eight 14-day cycles of intravenous nivolumab (240 mg). Oral exemestane 25 mg daily was started concomitant to nivolumab, and intramuscular triptorelin 3.75 mg every 28 days was started concomitant to chemotherapy. Endocrine therapy was maintained until surgery, which was performed 2 to 5 weeks after the last nivolumab dose. Adjuvant therapy was administered at physician’s discretion. Informed consent to participate in the study should be obtained from all participants. The study was been performed in accordance with the Declaration of Helsinki and approved by the competent ethical committee at each participating institution (Veneto Institute of Oncology IOV-IRCCS, Padova; Azienda USL-IRCCS, Reggio Emilia; University Hospital of Parma, Parma; Centro di Riferimento Oncologico di Aviano IRCCS, Aviano).
The primary endpoint was the rate of patients achieving a pCR by local pathology evaluation, defined as the absence of invasive cancer cells in breast and axilla (ypT0/is, ypN0)12. Among the 43 patients enrolled, 7 (16.3%) reached a pCR, with a rate of residual cancer burden class 0–I of 25.6%.
Gene-expression data from baseline tumor biopsies were available for all 43 patients enrolled in the study. Fresh-frozen baseline tumor biopsies, or alternatively FFPE baseline tumor biopsies if fresh-frozen tumor biopsy were not available, were reviewed by a pathologist for tumor tissue quality and quantity. All the biopsies contained at least 40% of tumor cells. Total RNA was extracted using the RNeasy Plus Mini kit (Qiagen) for fresh frozen tissue or RNeasy FFPE Kit (Qiagen) for FFPE tissue following the manufacturer’s instructions. RNA concentration, quality, and purity (260/280 ratio between 1.7 and 2.3; concentration ≥10 ng/μL) were assessed to ensure compliance with quality requirements. A minimum of approximately 100 ng of total RNA was then analyzed, according to manufacturer’s instructions, to measure the expression of 776 genes using the commercial Breast Cancer 360TM Panel on the nCounter Analysis System (NanoString Technologies).
Stromal TILs were evaluated on a hematoxylin- and eosin (H&E)–stained slide from diagnostic core biopsy according to the International Tumor-Infiltrating Lymphocytes Working Group recommendations22,23.
PD-1 and PD-L1 expression on T lymphocytes, macrophages and tumor cells was evaluated on FFPE biopsies using a multiplex immunofluorescence (mIF) approach. The Opal Polaris 7-Color IHC Detection Kit (Akoya Biosciences) was used to stain sequential 4 µm-thick FFPE tumor tissue sections with antibodies against CD68 (clone KP1, Dako), CD3 (clone F.7.2.38, Dako), PD-1 (clone EPR4877-2, Abcam), PD-L1 (clone E1L3N, Cell Signalling) and pan-cytokeratin (clone AE1/AE3, Dako). Multiplex stained slides were scanned using the Mantra Quantitative Pathology Workstation (Akoya Biosciences) and analyzed with InForm Image Analysis software (version 2.4.9, Akoya Biosciences). Cell densities (cells/mm2) in the intratumoral compartment (as identified by pan-cytokeratin) were calculated as the mean of all acquired fields on the same tissue slide (at least 20 fields at 20X magnification for each slide).
More detailed methods are reported in the previous publication12.
LETLOB neoadjuvant trial cohort
The LETLOB trial (NCT00422903) is an investigator-driven, multicenter, randomized phase-II trial, which included postmenopausal women with newly diagnosed, previously untreated, stage II-IIIA (T > 2 cm, N0-1) HR + /HER2- BC. Patients enrolled in the LETLOB trial were randomized to receive letrozole (2.5 mg/daily) plus lapatinib (1500 mg daily) or placebo for 6 months as neoadjuvant treatment. The primary end point was breast objective response rate assessed by ultrasonography24. Informed consent to participate in the study should be obtained from all participants. The study was been performed in accordance with the Declaration of Helsinki and approved by the competent ethical committee at each participating institution (Modena University Hospital, Modena; Ramazzini Hospital, Carpi; Azienda Ospedaliera Istituti Ospitalieri di Cremona, Cremona; Hospital of Piacenza, Piacenza; Azienda Ospedaliera Arcispedale S. Maria Nuova, IRCCS, Reggio Emilia; Ospedale “Infermi”, Rimini)
Azienda USL-IRCCS, Reggio Emilia; University Hospital of Parma, Parma; Centro di Riferimento Oncologico di Aviano IRCCS, Aviano).
Among the 92 enrolled patients, 43 received letrozole-lapatinib and 49 letrozole-placebo. Numerically similar clinical response rates (partial+complete response) were observed (70% for letrozole-lapatinib and 63% for letrozole-placebo). Mean Ki-67 expression decreased from 18.7% to 12.8% in the letrozole-lapatinib arm (p = 0.002) and from 19.2% to 12.8% in the letrozole-placebo arm (p < 0.001).
Gene-expression data from baseline tumor biopsies were available for 66 patients out of 92 patients enrolled in the study. As part of the original study protocol, RNA was extracted from pretreatment frozen core biopsies using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions and total RNA concentration and quality assessed. Samples that met quality requirements were then processed according to the Affymetrix GeneChip® 3′ IVT Express Kit user’s manual (Affymetrix, Santa Clara, CA), starting from 150 ng of total RNA for each sample16,25.
Probe-level data were normalized and converted to expression values using robust multiarray average (RMA) procedure. Quality control assessment was performed in R statistical environment using affy, affyQCReport, and affyPLM Bioconductor packages.
Stromal TILs were centrally evaluated on a hematoxylin- and eosin (H&E)–stained slide from diagnostic core biopsy according to the International Tumor-Infiltrating Lymphocytes Working Group recommendations22,23.
More detailed methods are reported in the previous publication24.
Gene-expression data analysis
Gene expression data from each of the two cohorts was separately analyzed. Gene expression data was log base2–transformed and normalized using housekeeping genes (ACTB, MRPL19, PSMC4, RPLP0, and SF3A1) and PAM50 subtype predictor was used to assign intrinsic subtype using nearest centroid procedure3. If the nearest centroid for a sample was Normal-like, second nearest centroid was selected.
In addition, previously published immune signatures associated with CD8 T-cells and Cytotoxic cells (as reported by BC360 Nanostring manufacturer’s information), and IFN-gamma15 were computed in each dataset separately.
In a previous exploratory analysis of the GeparNuevo trial, assessing the addition of durvalumab to neoadjuvant chemotherapy in triple-negative BC, 84 genes which were differentially expressed in pretreatment samples of patients according to pathologic complete response after neoadjuvant chemotherapy with durvalumab with an absolute log-fold-change>0.5 and p < 0.05 were identified20. This 84-gene signature, associated with response to both neoadjuvant chemotherapy and ICI in triple- also computed in each dataset separately.
These signatures were then rescaled on a 0 to 100 scale within each dataset and then jointly analyzed.
Statistical analysis
Subsequent statistical analyses were performed using R software v3.3.2 (R Development Core Team, Vienna, Austria) and IBM SPSS V28.0 software26.
Descriptive statistics were performed for patients’ demographics and clinicopathological characteristics. The Chi-squared test (χ2) test and Fisher’s exact test were used to study association between categorical variables, as appropriate. Kruskal–Wallis test and Mann–Whitney U test were used to study association between continuous variables and categorical variables, as appropriate. Spearman correlation was used to assess the association between two continuous variables.
All reported p values were two-sided and significance level was set at 5% (p < 0.05).
Data availability
Data that support the findings of this study are available from the corresponding author upon request, pending on formalization of a Data Transfer Agreement and reinforcing the compliance with the EU privacy law. Further information is available from the corresponding author upon request.
References
Nitz, U. A. et al. Endocrine therapy response and 21-gene expression assay for therapy guidance in HR+/HER2- early breast cancer. J. Clin. Oncol. 40, 2557–2567 (2022).
Loibl, S. et al. Early breast cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 35, 159–182 (2024).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009).
Cejalvo, J. M. et al. Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer. Cancer Treat. Rev. 67, 63–70 (2018).
Griguolo, G. et al. Gene-expression signatures to inform neoadjuvant treatment decision in HR+/HER2- breast cancer: available evidence and clinical implications. Cancer Treat. Rev. 102, 102323 (2022).
Bottosso, M. et al. Gene expression assays to tailor adjuvant endocrine therapy for HR+/HER2- breast cancer. Clin. Cancer Res. 30, 2884–2894 (2024).
Bertucci, F., Finetti, P., Goncalves, A. & Birnbaum, D. The therapeutic response of ER+/HER2- breast cancers differs according to the molecular Basal or Luminal subtype. NPJ Breast Cancer 6, 8 (2020).
Prat, A. et al. A PAM50-based chemoendocrine score for hormone receptor-positive breast cancer with an intermediate risk of relapse. Clin. Cancer Res. 23, 3035–3044 (2017).
Schmid, P. et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med 386, 556–567 (2022).
Cardoso, F. et al. Pembrolizumab and chemotherapy in high-risk, early-stage, ER+/HER2− breast cancer: a randomized phase 3 trial. Nat. Med 31, 442–448 (2025).
Loi, S. et al. Neoadjuvant nivolumab and chemotherapy in early estrogen receptor-positive breast cancer: a randomized phase 3 trial. Nat. Med 31, 433–441 (2025).
Dieci, M. V. et al. Neoadjuvant chemotherapy and immunotherapy in luminal B-like breast cancer: results of the phase II GIADA trial. Clin. Cancer Res 28, 308–317 (2022).
Criscitiello, C. et al. Tumor-infiltrating lymphocytes (TILs) in ER+/HER2− breast cancer. Breast Cancer Res. Treat. 183, 347–354 (2020).
Fujimoto, Y. et al. Prognostic significance of tumor-infiltrating lymphocytes may differ depending on Ki67 expression levels in estrogen receptor-positive/HER2-negative operated breast cancers. Breast Cancer 26, 738–747 (2019).
Ayers, M. et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 127, 2930–2940 (2017).
Griguolo, G. et al. Immune microenvironment and intrinsic subtyping in hormone receptor-positive/HER2-negative breast cancer. NPJ Breast Cancer 7. https://doi.org/10.1038/s41523-021-00223-x (2021).
Huppert, L. A. et al. Pathologic complete response (pCR) rates for patients with HR+/HER2− high-risk, early-stage breast cancer (EBC) by clinical and molecular features in the phase II I-SPY2 clinical trial. Ann. Oncol. 36, 172–184 (2025).
Nanda, R. et al. Effect of pembrolizumab plus neoadjuvant chemotherapy on pathologic complete response in women with early-stage breast cancer: an analysis of the ongoing phase 2 adaptively randomized I-SPY2 trial. JAMA Oncol. 6, 676–684 (2020).
Mittempergher, L. et al. The ImPrint immune signature to identify patients with high-risk early breast cancer who may benefit from PD1 checkpoint inhibition in I-SPY2. J. Clin. Oncol. 40, 514–514 (2022).
Sinn, B. V. et al. Immune-related gene expression predicts response to neoadjuvant chemotherapy but not additional benefit from PD-L1 inhibition in women with early triple-negative breast cancer. Clin. Cancer Res. 27, 2584–2591 (2021).
Ayers, M. et al. Relationship between immune gene signatures and clinical response to PD-1 blockade with pembrolizumab (MK-3475) in patients with advanced solid tumors. J. Immunother. Cancer 3, P80 (2015).
Hendry, S. et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the international immunooncology biomarkers working group: part 1: assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv. Anat. Pathol. 24, 235–251 (2017).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 26, 259–271 (2015).
Guarneri, V. et al. Double-blind, placebo-controlled, multicenter, randomized, phase IIB neoadjuvant study of letrozole-lapatinib in postmenopausal hormone receptor–positive, human epidermal growth factor receptor 2–negative, operable breast cancer. J. Clin. Oncol. 32, 1050–1057 (2014).
Guarneri, V. et al. Double-blind, placebo-controlled, multicenter, randomized, phase IIB neoadjuvant study of letrozole-lapatinib in postmenopausal hormone receptor-positive, human epidermal growth factor receptor 2-negative, operable breast cancer. J. Clin. Oncol. 32, 1050–1057 (2014).
The R foundation. R: The R project for statistical computing. Available at https://www.r-project. org n.d.
Acknowledgements
This work was supported by University of Padova, Department of Surgery, Oncology and Gastroenterology DOR 2023-2024 (to V.G., M.V.D., G.G., F.M.; grant number not applicable), Ricerca Corrente funding from the Italian Ministry of Health (grant number not applicable). Fondazione AIRC under 5 per mille 2019 (ID. 22759 program—group leader VG) and under IG 27152 to M.V.D.
Author information
Authors and Affiliations
Contributions
Study design: G.G., M.V.D. and V.G.; Acquisition of clinical data: G.G., M.B., F.M., D.G.G., A.M., S.S., G.M.V., T.G., R.C., F.P., K.G., M.V.D., and V.G.; Translational Analyses, data analysis and interpretation: G.G., M.B., L.P., R.C., S.B., E.T., A.T., F.S., A.P., and M.V.D.; Manuscript drafting: M.B. and G.G.; Manuscript revision and final approval of manuscript: all authors.
Corresponding author
Ethics declarations
Competing interests
G.G. reports fees for advisory role from Gilead, Seagen, Menarini, personal fees as an invited speaker from Eli Lilly, Novartis, MSD. M.B. reports travel support from Eli Lilly. F.M. reports personal fees from Roche, Novartis, Pfizer, Seagen, Menarini, MSD, Gilead, Astrazeneca. A.M. report consulting or advisory role from Eli Lilly, Eisai Europe, Daiichi Sankyo/Astra Zeneca, Novartis, research funding from Eli Lilly and Roche, travel/accommodations from Pfizer. S.S. reports the following: Novartis (speaker honoraria, advisory board, travel and expense Reimbursement for congress), Daiichi-Sankyo (speaker honoraria, tutorship, advisory board, travel and expense reimbursement for congress), Roche (travel and expense reimbursement for congress), Seagen, MSD and AstraZeneka (speaker honoraria and advisory board), Gentili and Mundipharma (speaker honoraria). T.G. reports personal fees from Novartis and Eli Lilly. F.P. reports, outside the submitted work, the following: consultancy/advisory board for Daichii Sankyo, Novartis, Pfizer, Roche; honoraria as a speaker from Novartis, Lilly, MSD, Pfizer. A.P. reports advisory and consulting fees from AstraZeneca, Roche, Pfizer, Novartis, Daiichi Sankyo, Ona Therapeutics, and Peptomyc, lecture fees from AstraZeneca, Roche, Novartis, and Daiichi Sankyo, institutional financial interests from AstraZeneca, Novartis, Roche, and Daiichi Sankyo; stockholder and employee of Reveal Genomics; patents filed PCT/EP2016/080056, PCT/EP2022/086493, PCT/EP2023/060810, EP23382703, and EP23383369. M.V.D. reports personal fees for consultancy/advisory role from: Eli Lilly, Pfizer, Novartis, Seagen, Gilead, MSD, Exact Sciences, AstraZeneca, Roche, Daiichi Sankyo, Roche. V.G. reports personal fees for advisory board participation for AstraZeneca, Daiichi Sankyo, Eli Lilly, Gilead, MSD, Novartis, Pfizer, Pierre Fabre, Roche, Menarini Stemline, Exact Sciences, personal fees as an invited speaker for AstraZeneca, Daiichi Sankyo, Eli Lilly, Exact Sciences, Gilead, GSK, Novartis, Roche, Zentiva, Menarini Stemline, personal fees for expert testimony for Eli Lilly, patents for HER2DX (Institution). All remaining authors have declared no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Griguolo, G., Bottosso, M., Paré, L. et al. Basal-like HR + /HER2- breast cancers show higher tumor-infiltrating lymphocytes and immune signatures with potential therapeutic implications. npj Breast Cancer 12, 23 (2026). https://doi.org/10.1038/s41523-025-00886-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41523-025-00886-w