Abstract
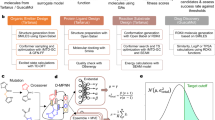
In this paper, we propose a novel flexible optimization pipeline for determining the optimal adsorption sites, named AUGUR (Aware of Uncertainty Graph Unit Regression). Our model combines graph neural networks and Gaussian processes to create a flexible, efficient, symmetry-aware, translation, and rotation-invariant predictor with inbuilt uncertainty quantification. This predictor is then used as a surrogate for a data-efficient Bayesian Optimization scheme to determine the optimal adsorption positions. This pipeline determines the optimal position of large and complicated clusters with far fewer iterations than current state-of-the-art approaches. Further, it does not rely on hand-crafted features and can be seamlessly employed on any molecule without any alterations. Additionally, the pooling properties of graphs allow for the processing of molecules of different sizes by the same model. This allows the energy prediction of computationally demanding systems by a model trained on comparatively smaller and less expensive ones.
Similar content being viewed by others
Introduction
Novel, functional structures at the nanoscale could be crucial for transforming a broad spectrum of economically significant processes into greener and more sustainable solutions. For instance, nanostructured materials hold the potential to significantly enhance the cost-effectiveness of fuel-cell devices1, enable the creation of highly efficient quantum-dot LEDs2, and pave the way for generating atom-precise efficient nanocatalysts for studying novel catalytic pathways in electrochemical applications3,4.
As performance is highly dependent on specific structural characteristics which often can not easily be resolved in lab experiments, computational chemistry - most often by using Density Functional Theory (DFT) based approaches - can be used to generate in-silico insights. Typical questions range from elucidating which feature of a given nanoparticle might improve catalytic performance to mechanistic explanations for key synthesis procedures, allowing tailored experiments to drive up experimental yields for optimal structures.
Commonly, these questions are associated with finding energetically favorable configurations for the potential energy surface (PES) of a system, which is a property relevant to solving a wide range of problems in computational chemistry.
The established methodology allows finding “docking” mechanisms between small molecules and large biomolecules, which is relevant for drug development5. Additionally, a large area of research revolves around the sensing of harmful gases by novel nanomaterials chosen according to their strength of interactions. In the field of catalysis, identifying and maximizing relevant catalytic sites with the lowest number of atoms results in highly efficient and financially viable catalysts6.
For the latter, the primary approach starts by finding symmetry points of a surface crystal plane and sampling a small area of the configuration space around them7 to find a minimal energy configuration. All system constituents are usually kept rigid in this approach, and only the final configurational pick is subject to a relaxation procedure and further study. Over time, several physics-based optimization techniques have evolved to allow for a more elaborate study of the PES moving beyond simple sampling of enumerated configurations: the Nudged Elastic Band (NEB) method allows the description of catalysis pathways8. With Minima-Hopping and genetic algorithms, new stable material conformations can be explored9,10, and metadynamics could be used to examine the thermodynamics of a given system11,12. Despite these methodological improvements, the high number of necessary energy evaluations has relegated those to niche and specialist applications. At the same time, the original sampling-based workflows for configurational search are still in everyday use and can be performed in a highly automated fashion13,14. However, when applied to novel systems incorporating irregularly shaped nanostructures or more complex adsorbents, exhaustive sampling quickly reaches the computational limits of the average project. Here, data-driven machine learning techniques such as neural networks have been highly utilized15 and could be used to describe the PES and subsequently allow for more efficient exploration. While early research showed that this approach could successfully reproduce even complex PES16,17, the practical application requires that this can be achieved without exhaustively sampling the PES of the system under examination for data generation.
One approach is using a large, pre-trained model describing the system and using that to significantly speed up an exhaustive sampling and relaxation procedure, as has been done to model adsorption dynamics18,19. However, this relies on the availability of a large-scale dataset to create a model with suitable generalization power towards the research problem. Without such prerequisites, one can also iteratively build up an uncertainty-quantifying model for the PES of a specific system using techniques such as active learning (AL) or Bayesian optimization (BO). The “classic” approach to this problem would be a Gaussian-process (GP) based regression model for the PES. The inputs would be features extracted from the atomistic structure20,21, with newer research suggesting the usage of end-to-end-trained graph-neural-network models22. These kinds of machine-learning models have been used to find stable crystal structures23,24,25, learn adsorbate-surface PES26 and have been integrated into techniques like NEB27,28 or minima-hopping29 resulting in significant computational speedup.
The basic problem of finding energetically minimal configurations for a variety of systems using BO has been extensively studied under the BOSS-handle using problem-specific features and Gaussian process-based PES surrogates. Starting by demonstrating that energetical minimal molecular structures can be found by optimizing a model based on dihedral angles30, the method was further demonstrated for adsorption problems by finding the optimal placement of large, rigid molecules on inorganic surfaces31,32 and has been refined to determine optimal adsorption structures for partly-flexible molecules on gold-clusters33. They, however, rely on hand-crafted feature extraction functions, which can restrict the potential expressibility of the model and require a high level of prior physical understanding that might not be present in more complex systems. Further, this approach isn’t transferable as it relies on a fixed, molecule-dependent size of input features.
Our method encodes the cluster-adsorbate system as a graph. This is processed by a state-of-the-art Graph Neural Network (GNN), which allows the representation to be symmetry, rotation, and translation invariant. Given the pooling properties of the graph, this output has the same dimensionality independently of the molecule used for input. The output of the GNN is used as input to a Gaussian Processes (GP) model which creates the final predictions along with quantification of their uncertainty. Finally, a Bayesian Optimization (BO) scheme produces suggestions for optimal adsorption sites.
We demonstrate the robustness of the AUGUR framework on four systems. The first is the family of the chini clusters that have been exhaustively studied as a precursor to generating atom-precise nanoclusters using Molecular Organic Framework as a template in our previous study3. The atom-precise nanoclusters are crucial in developing the next generation of highly tailored, efficient, and economically viable nanocatalysts. The DFT simulations of Zn2+ - Chini cluster interactions found in literature allow us to know, a priori, the energetic behavior of all chemically distinct sites. This, in turn, allows the validation of our algorithm. Second, we selected ZnO clusters that are well-known experimentally to be used in semiconductor companies for chips, sensors, and electrodes due to their observed high catalytic activity and stability34. In our study, the focus lies on the gas adsorption capability of the (ZnO)78 cluster, as it is one of the “magic clusters”35. However, due to the large size of the cluster, the conventional approach of exhaustive DFT investigation, covering the entire surface of the cluster, is computationally infeasible. The third and fourth use cases involve extended 2D surfaces with single and multiple defects, respectively. The third case investigates a hydrogen-deficient Covalent Organic Framework (COF), TpBD-FF, whose reactivity arises from this structural deficiency. Examining its interactions with Zn ions from the electrolyte provides valuable insights into its potential as an anode coating material in zinc-ion batteries36. Our fourth case study explores a multi-atom substituted Silicene, a 2D allotrope of Silicon whose enhanced electronic structure properties make it a promising candidate for many catalytic applications37,38. In this work, we specifically focus on formaldehyde adsorption on the defective Silicene layer.
The AUGUR pipeline has proven itself robust, transferable, and highly efficient in all case studies, identifying the optimal sites with approximately ten simulation runs and without the need for hand-crafted features.
Results
To show the generalizability of the method, we investigate two different adsorption scenarios, featuring both atomic and molecular adsorbates, along with four nanosystem adsorbents of varying complexity. The adsorbate distance was chosen based on the explicit literature survey of the optimal interaction distance between different atomic species. Nevertheless, AUGUR offers the functionality of determining these distances as the sum of the van der Waals radii. This is a less efficient approach as it places adsorbates further from the cluster/surface than necessary, increasing DFT times. It however, adds another level of abstruction allowing AUGUR to be used in an even more problem-agnostic manner.
Cluster adsorption optimization
Chini clusters
Chini clusters are a unique class of organometallic compounds known for their unusual bonding and electronic properties. These clusters serve as precursors for synthesizing atomically precise nanoclusters, with their model structures illustrated in Fig. 1. Understanding the interaction between Chini clusters and Zn2+ ions is crucial for unraveling the encapsulation process of platinum nanoparticles (Pt NPs) within the Zn-based ZIF-8 Metal-Organic Framework (MOF)3 template. The precise understanding and control of this interaction will aid the creation of highly efficient and specialized electrocatalysts that minimize the usage of rare earth elements.
For generating suggestions for the optimal adsorption positions of Zn ions on the Chini clusters, point generation was guided by the following physical principles: The Zn atom was positioned on the surface of a sphere centered on either Pt or O atoms. Carbon atoms were excluded as potential adsorption sites since they are unavailable for interaction on the cluster’s surface. A mixture of initial simulations and physical intuition determined the sphere’s radius. It must be noted that this constraint can err on the high side, as the only effect will be an increase in the computational time, with the final energy remaining constant. After evaluating these points, our pipeline suggested the optimal adsorption sites, which were then simulated using DFT.
The interaction energies at these suggested sites were compared with those obtained from Monte Carlo (MC) sampling. As shown in Table 1, the lowest interaction energy predicted by AUGUR is consistently lower than that predicted by the MC approach, a trend that becomes more pronounced with increasing cluster size-from an improvement of 8.73 % in Pt3 to 142.62 % in Pt9. This demonstrates that our framework is more robust in finding the global minima than the random MC sampling approach, ultimately saving both time and resources.
In addition to the optimization process, the high level of symmetry allows for fairly accurate energy predictions. This was evaluated using Leave One Out (LOO) cross-validation. Although the uncertainty was relatively high due to the limited number of points required, the Mean Square Error (MSE), as shown in Table 2, was seen to be remarkably low. A more detailed representation of the LOO can be seen in the SI.
Given the accuracy showcased in Table 2, our model can be leveraged to reconstruct the entire energy surface and quantify uncertainties in unexplored regions. This approach offers a more holistic understanding of the system as compared to the traditional trial-and-error sampling, which lacks an underlying model for further system interpretation.
Prior literature knowledge of the electronic structure of Chini clusters allows us to verify the results. These clusters have three distinct chemical sites: the “Top,” “Bridged,” and “Terminal”. Our study shows that the “Top” is the most energetically favorable for Zn ion binding. This is due to the formation of the strong Zn-Pt bonds at the exposed Pt atoms in the outer layer of the cluster, as concluded by our previous study3. Additionally, the “Bridged” and the “Terminal” positions show considerably less strong interaction energies due to the carbonyl ligands inhibiting the direct Metal-Metal (M-M) bonding3. AUGUR successfully recreates these findings, accurately identifying the optimal and relative energetics of Zn-chini cluster interactions.
In Fig. 2, the energy surface and optimal site for Pt3 are presented and verified with the predictions. Of note is that although the predictions are fairly accurate, the uncertainty is relatively high. This can be attributed to the low number of points, as well as to the different distances of Zn ion placement in the training and prediction sets. For a more detailed view of the energy surface and standard deviation, refer to the SI.
In Fig. 3, the energy surface and optimal site for Pt6 are presented. The uncertainty is equivalent to that in Fig. 2 despite the larger size. This is because Pt6 exhibits a strong symmetry, which reduces the number of unique sites to those of Pt3. For a more detailed visualization of the energy surface and standard deviation, refer to the SI.
The Pt9 cluster introduces two additional distinct sites due to the existence of the middle layer. These are the “Bridged Middle” and “Terminal Middle”. According to the prior analyses3, these sites are expected to be less energetically favorable but follow the same energy relationships as those in the upper and lower layers. In the SI, a detailed view of the energy surface and standard deviation is provided. Of note is that this prediction was generated with only one simulation from Pt9 included in the training set, alongside Pt3 and Pt6. This finding is of potentially high importance as it suggests that our model could be trained on comparatively inexpensive simulations supplemented by a few strategic points from the more computationally intensive cases to achieve physically consistent and time-efficient results. These models could then be deployed for the prediction and optimization of larger structures of the same family and related systems with higher nuclearity, which would otherwise be computationally infeasible to simulate as seen in Fig. 4.
Nevertheless, it is important to note that the Chini clusters system is relatively simple, and intuition alone would have converged to the optimal site in a reasonable number of iterations. Therefore, our pipeline was subsequently tested on a much more complex system, lacking obvious symmetries.
Zinc Oxide Cluster
Our second case study involves the Zinc oxide ((ZnO)78) cluster depicted in Fig. 5, with carbon monoxide (CO) as the adsorbate molecule. ZnO clusters are used experimentally and industrially in semiconductor companies for chips, sensors, and electrodes due to their observed high catalytic activity and stability34. Our study aims to aid the exploration of the gas adsorption potential of the large Zn oxide clusters, which would otherwise be very computationally expensive to pursue. The (ZnO)78 was mainly selected because it is one of the “magic clusters”39, yet remains relatively unexplored in the literature. Given the complexity and asymmetry of such an extensive system, manually identifying all the chemically distinct adsorption sites on the potential energy surface (PES) to begin the exploration is impractical. Through our investigation, we discovered two distinct sites in what appeared to be nearly identical locations. However, the adsorption energies varied significantly between these sites, with one being the optimal adsorption position and the other showing only half the absolute adsorption energy.
The point generation in this study was done by positioning the carbon atom of the adsorbate on the surface of a sphere centered on a randomly selected atom within the cluster, with a radius of 1.40 Å. Subsequently, the oxygen atom of the adsorbate was placed on the surface of a sphere centered on the carbon atom, with a radius of 1.12 Å, corresponding to a triple bond between the C and O atoms of carbon monoxide. The placement was further constrained to maintain a minimum distance of 1.40 Å between the oxygen atom and the Zinc oxide cluster so that it does not coincide with the cluster atoms. Additionally, the carbon atom was positioned first as it shows a higher chemical affinity for the cluster. In this way, we ensure that our method does not preclude placements of the adsorbate where the O atom is closer to the cluster, but makes it less likely.
We present the comparison of energy optimization results obtained using both Monte Carlo (MC) sampling and the AUGUR framework in Table 3. In total, 19 samples were evaluated with the MC approach, compared to 13 with AUGUR. Despite the lower number of samples, the AUGUR framework consistently outperformed the MC method in both energy optimization and data efficiency. To ensure that the optimum identified by AUGUR is indeed the global minimum, approximately 70 additional simulations were conducted. After thoroughly investigating the structure, we are confident that the identified optimum is the global minimum.
An additional advantage of this approach is its ability to generate data points that train an accurate model in the most data-efficient manner. To demonstrate this, we present the cross-validation results from both approaches in 3. Each AUGUR step involved two suggestions: one focused on extreme exploration (with a tradeoff of 100) and the other on extreme exploitation (with a tradeoff of 0.5). This method could be further refined by gradually decreasing the tradeoff and generating only one suggestion per step. However, the combined approach enabled rapid convergence-not in terms of the total number of simulations but in terms of the number of suggestion steps-by allowing the two extremes to be evaluated in parallel.
In Fig. 6, we show the predicted energy surfaces and associated uncertainties of the investigated cluster by predictions made at 5000 points. The prediction of those suggested points required approximately 150 seconds. In addition, the average simulation duration for one suggestion of this case study is approximately two days. Therefore, performing an exhaustive energy surface investigation using traditional means would have taken multiple years.
For a more detailed view of the energy surface and standard deviation, refer to the SI. This case study, more than anything, highlights the advantages of the AUGUR framework. Our pipeline not only determines the best energy site with minimal data cost, it also gathers points efficiently for the model’s training which can be seamlessly used to gain physical insight that would have otherwise taken years to achieve.
Surface adsorption optimization
In this section, we investigated 2D surface adsorption. The molecule’s placement was adjusted to prevent unphysical configurations, such as penetrating the surface, with periodic boundaries accounted for using the minimum image convention40. This new distance calculation was used to both calculate the edge attributes of the graphs, as well as assure that the adsorbate was placed within the allowed distance to the cluster, accounting for periodicity. Geometric constraints, avoiding unphysical overlaps or piercing of the surface, were further implemented.
Defective COF surface
For our third case study, we examined the TpBD-FF Covalent Organic Framework (COF), a two-dimensional porous periodic system known for its use as a coating material for zinc-ion battery (ZIB) anodes. The structure of TpBD-FF is illustrated in Fig. 7 a. TpBD-FF enhances cell efficiency by promoting the formation of well-defined ion transport channels36. Our goal is to understand this by studying the interaction of electrolytic Zn2+ ions with a single defective layer of TpBD-FF. We aim to identify the optimal adsorption site and its strength, which is essential for evaluating which specific functional groups in the COF layer influence its protective role in ZIB performance.
This figure depicts the relaxed structure of the (a) Bare TpBD-FF COF and (b) H-deficient TpBD-FF COF, viewed along the z-axis. The pink encircling in b shows the position where the H-deficiency defect was introduced. The color scheme used is as follows: O (red); C (light grey); N (Blue), F(Fluorine), H(light pink).
To test the robustness of the AUGUR framework in defective periodic systems, we introduced a hydrogen deficiency defect, a commonly observed imperfection in COFs41. Specifically, a hydrogen atom was removed from one of the chemically reactive N-H bonds of TpBD-FF, as illustrated in Fig. 7b. Introducing such a defect not only mimics realistic conditions but also allows us to test the more accurate applicability of the TpBD-FF COF to be used as an anode coating.
To systematically determine the optimal interacting site, Zn2+ ions were initially placed at three random positions on the TpBD-FF surface. The point generation process was performed similarly to the case study described in Section “Cluster adsorption optimization”, where a single-atom adsorbate was considered, too. Specifically, Zn2+ ions were positioned at distances ranging from 1.6–2.4 Å from all the surface available atomic species (H, O, and F) within TpBD-FF, ensuring sufficient sampling of the potential energy surface. This range was considered from the various existing experimental and computational distances - Zn-H42, Zn-O43, Zn-F44) reported in the existing literature.
The energy optimization results comparing the Monte Carlo (MC) sampling approach and the AUGUR framework are summarized in Table 4. A total of 10 candidate configurations were evaluated using MC sampling, whereas the AUGUR framework converged in just 1 iteration. The most optimal binding site has the lowest energy −8.00 eV as compared to −6.44 eV in MC samples. Despite the significantly reduced number of samples, AUGUR consistently outperformed MC sampling in terms of energy optimization and computational efficiency.
To validate that the site identified by AUGUR corresponds to the global energy minimum, we exhaustively evaluated all unique adsorption sites on the TpBD-FF surface. This exhaustive search, as shown in Fig. 8, confirmed that AUGUR’s prediction aligns with the true global minimum, further demonstrating the robustness and reliability of the AUGUR framework in handling adsorption problems within complex 2D periodic systems. For a more detailed view of the energy surface and standard deviation, refer to the SI.
Defective silicene
Our final case study focuses on Silicene, a 2D periodic material, visualized in Fig. 9a), that exemplifies the broad applicability and robustness of the AUGUR pipeline. Silicene features a buckled structure arising from the sp2-sp3 hybridization of silicon atoms. This structural corrugation endows Silicene with unique electronic properties and an active surface, making it a promising candidate for various chemical applications37,38. Additionally, its defect engineering has been extensively studied to enhance chemical reactivity for specific purposes45. These attributes render Silicene an ideal test case for validating the effectiveness of AUGUR in handling periodic systems, especially with multiple defects.
To demonstrate AUGUR’s capability in such challenging use case scenarios, we introduced five distinct point defects into the Silicene lattice, all of which have been previously characterized separately in the literature46,47,48,49,50. Specifically, we replaced five randomly selected silicon atoms with boron (B), aluminum (Al), carbon (C), nitrogen (N), and phosphorous (P). This defective Silicene structure was then employed as an adsorbent substrate to evaluate whether AUGUR could efficiently and accurately converge to the most favorable adsorption site for an incoming adsorbent molecule.
We selected formaldehyde (CH2O) as the adsorbate for two key reasons. First, formaldehyde is a well-known carcinogen, and studying its interaction with defective Silicene could offer insights into its potential use as a sensor for its atmospheric or biological detection51. Second, formaldehyde serves as a sufficiently complex test case, with its four constituent atoms allowing us to assess AUGUR’s efficacy in cases where higher-dimensional adsorbates are involved.
To address the complexity of the adsorption process, we implemented an enhanced placement strategy that positioned the entire adsorbate molecule in a single step, followed by a random rotational adjustment. This approach allowed for systematic exploration of adsorption configurations while maintaining computational efficiency. Furthermore, the placement methodology prioritized the aldehydic oxygen atom in formaldehyde, ensuring its alignment towards the defective Silicene surface, as it is the most reactive functional group in formaldehyde. To achieve this, the oxygen atom was initially positioned near a randomly selected Silicene surface atom at specific distances informed by the experimentally and computationally established bond lengths in the literature cited. The selected placement distance ranges were: O-Si (1.57–1.73 Å)52,53, O-B (1.28–1.46 Å)54,55, O-C (1.13–1.48 Å)56,57, O-N (1.1–1.44 Å)58, O-P (1.43–1.52 Å)59,60, and O-Al (1.68–2.1 Å)61,62. While these ranges were considered for completeness, placement was consistently performed at the lower bound of each range to ensure uniformity across configurations. Subsequently, the formaldehyde molecule was rotated randomly along its three principal axes. This rotation was carefully designed to preserve the proximity of the oxygen atom to the substrate, ensuring that it remained the closest point of interaction with the defective surface.
As can be seen from Table 5, AUGUR significantly outperformed the MC benchmark, with the lowest interaction energy being −2.19 eV as compared to −1.80 with MC sampling.
Lastly, the energy surface along with the optimal adsorption position are presented in Fig. 10. For more details about the energy surface and standard deviation, refer to SI.
Discussion
In summary, we have presented a novel deep kernel learning framework for automatically determining the optimal adsorption sites on molecules, minimizing the number of DFT simulations required. It seamlessly combines graph neural networks and Gaussian processes. In this way, we achieve rotation, symmetry, and translation invariance for the inputs of the Gaussian processes. Additionally, because of the properties of graphs, we also allow for the same model to be used across different molecules regardless of their size. This alleviates the need for crude techniques such as padding or hand-crafted features that require a high level of physical intuition. Further, this allows for significant dimensionality reduction to the inputs of the Gaussian processes, which famously scale poorly with increasing feature dimensions. This composite model will enable us to fully investigate the energetic structure of the molecule as well as selectively refine areas of high uncertainty if deemed necessary. The model is then used as the surrogate to a Bayesian Optimization scheme that iteratively refined both the model and the suggested sites for optimal adsorption till it converges to the global optimum.
It is shown that this algorithm performs robustly and efficiently, determining both the optimal sites as well as predicting the energy surface with a limited number of data points. The natural next step is to deploy this algorithm for cases of highly complex adsorbents and leverage AUGUR to further our chemical understanding of hitherto computationally prohibitive systems.
Methods
AUGUR pipeline
Our pipeline consists of four major components, also visualized in Fig. 11:
-
A graph neural network that is responsible for extracting a meaningful, rotation, translation, and symmetry invariant representation of the molecule. The output dimension of the graph is fixed regardless of the molecule’s size and, therefore, can be used across various molecules.
-
A Gaussian Process that receives as input the output of the graph and predicts the system’s interaction energy along with the corresponding uncertainty in the prediction
-
A Bayesian Optimizer that uses the above two models as a combined surrogate and generates suggestions for adsorption positions. These are simulated by the simulation model.
-
A Density Functional Theory model that provides a physically accurate but computationally expensive evaluation of the interaction energy of the suggestions.
The top line is the optimization pipeline, from left to right, training the GNNs and the GPs, using them as surrogates for BO, evaluating the BO suggestions with DFT, adding the new results into the dataset, and repeating them. The bottom line is the point generation for BO, from left to right, defines the cluster, place the first adsorption atom at a physically consistent distance and outside the convex hull of the molecule, and repeats this process with all atoms of the adsorbant molecule.
It must be noted that the exact model architecture will be less relevant with the number of samples going to infinity. Since we are operating on a limited dataset, however, we need to decide on the most efficient architecture parameters. There are some broad guidelines that can be followed. The number of convolution layers for example, can be chosen to roughly correspond to the maximum number of n-hop neighbors that affect each atom’s interaction. The GP kernel can be chosen to reflect the perceived smoothness or discontinuity of the interaction energy surface, and if we standardize the data, the zero mean can be chosen as the prior mean. These guidelines, however, are not guaranteed to yield optimal results, not to mention the lack of specific knowledge for these specifications. To this end, we perform a model parameter sweep and choose the one that yields the lowest test set MSE. Lastly, the pipeline provides a prediction for the overall interaction energy surface and its corresponding uncertainty. The combination of those will allow the user to decide on an individual basis whether the predictions are sufficiently accurate for the desired application.
Density Functional Theory
Density Functional Theory (DFT) is an approximation method used to solve the Schrödinger equation for many-body systems.
The total energy (functional of density) of the system is given by:
where Ts[ρ] is the kinetic energy of the non-interacting electrons, EH[ρ] is the Hartree energy representing the electron-electron repulsion, Eext[ρ] is the external potential energy from the nuclei, and Exc[ρ] is the exchange-correlation energy that includes all complex quantum mechanical effects.
The variational principle used to determine the ground-state energy is given by:
Density Functional Theory (DFT) is particularly valued for its computational efficiency as, unlike traditional ab-initio methods, DFT scales more favorably at N3. In this context, N represents the system size, encompassing the number of atoms, electrons, or basis functions. Consequently, if the system size doubles, it increases only by a factor of 8. This more favorable scaling allows DFT to perform calculations faster and handle larger systems more effectively than traditional ab-initio methods like Hartree Fock (HF). Furthermore, DFT inherently accounts for electron correlation effects, which are often neglected by HF. This capability makes DFT especially useful for systems where electron correlation is significant, such as in transition metal chemistry, which is central to many chemical reactions and the use cases in our study.
Despite the advantages, DFT does have limitations, particularly in terms of system size. Although DFT simplifies the many-body problem of N electrons to a dependency on the electronic density (reducing the complexity to three spatial coordinates), it is generally constrained to handling systems with a few hundred atoms. The computational effort still scales approximately as N3, making the study of electronic structure properties of large systems resource-intensive and time-consuming.
To mitigate these limitations, our study leverages AUGUR, a framework that guides the experimental process using Bayesian Optimization (BO) enhanced by graph-based Gaussian Processes. This approach reduces the number of required DFT simulations, significantly lowering the computational burden.
Bayesian Optimization
Bayesian Optimization (BO) is an optimization algorithm that has proven itself to be both data efficient and accurate even in non-convex/concave optimization problems. In principle, let us consider a costly optimization problem, such as one relying on expensive simulations or arduous experimental work for the evaluation of the objective function. The natural solution to cases such as these is the training of a surrogate model that will provide sufficient accuracy at a fraction of the required time. Nevertheless, data-driven models typically require a lot of data to be trained sufficiently, which in turn invalidates the main motivation of the surrogate model, i.e., the minimization of the requirement of the time-consuming part of the pipeline. Bayesian optimization is particularly suited to this kind of problem because it simultaneously trains a surrogate model efficiently and identifies hopeful query points. It relies on two components. The first is a stochastic predictor, i.e., a predictor that can provide uncertainty quantification, typically Gaussian Processes (GP), Bayesian Neural Networks (BNN), or others. The second is a function that evaluates each possible point on a) how optimal it is and b) how much new information it injects into the model. This function shall be referred to as the acquisition function. For the remainder of the chapter, we will describe a minimization process, but everything can be applied without loss of generality to any optimization problem. In the present work, we seek to find the position of adsorption that will result in the lowest possible energy. To this end, we can generate a number of random positions and evaluate their respective energies using the uncertainty-aware predictor. The energy value is an indicator of the site’s optimality. The standard deviation reflects how uncertain the prediction is. Choosing to focus on the points of high standard deviation will lead to the most efficient injection of new information and, therefore, the fastest training of the surrogate model. This is called exploration and is typically the focus of the first queries of the ground truth process. As the surrogate improves, the focus shifts to determining the optimal points, and therefore, the choice is increasingly being determined by the prediction and less by the standard deviation. To accommodate both exploration and exploitation efficiently, we have simulated a point with tradeoff 0.5 and a point with tradeoff 100. The first point prioritized areas of strong interactions and the second areas of high uncertainty. This phase is called exploitation. A more detailed derivation can be found in SI.
Note that there are multiple, much more efficient ways of optimizing the acquisition functions, including gradient-based and evolutionary algorithms. Nevertheless, the complexity of the constraints of this problem made their application non-trivial. In comparison, random point generation proved very efficient, especially given that the deep kernel method allows for the fast and parallel processing of thousands of points in mere seconds. More information about how the points were generated can be found in Section “Point generation”.
Point generation
The convergence of Bayesian Optimization can be significantly sped up if we introduce a layer of physical understanding of the problem. The simplest way is to generate a number of points (in our case, 10,000) that adhere to a set of constraints. A random atom of the cluster is chosen. The adsorbing atom is placed on the surface of a sphere with a radius determined by the physical limits of the interaction distance between the two atom types. Then, the adsorbate is tested for being placed inside the cluster or outside. This is done by performing a Delaunay tessellation on the cluster coordinates and determining the location of the adsorbent with respect to the resultant convex hull. If the adsorbent is found inside the hull, it is discarded, and the process starts anew. If this constraint is not violated, the adsorbent distance to the remaining cluster atoms is determined. If it was not positioned closer than the acceptable limits for any atom, then the new adsorbent position is accepted into the sample set and evaluated through the acquisition function. Otherwise, it is discarded, and the process starts anew. The sample generation process can be summarized as follows:
-
1.
Choose a random atom of the cluster, provided adsorption is physically possible on it.
-
2.
Place the adsorbent on the surface of a sphere with a radius determined by physical knowledge of the maximum and minimum interaction distance of these two atoms.
-
3.
Test if the adsorbent falls inside the convex hull of the cluster or goes through the adsorbate surface.
-
4.
If the above constraint is not violated, test if the adsorbent is closer than a distance of tolerance to the other atoms.
-
5.
If the above constraint is not violated, the sample is admitted into the list of viable samples.
If the adsorbent consists of more than one atom, we place every subsequent atom on a sphere with a radius chosen based on the expected bond length and repeat steps 3 and 4 until all the adsorbent atoms have been placed. Then, the total system is accepted into the list of viable samples. A concise flowchart of the process can be found in Fig. 11.
Gaussian processes
In the current project, the surrogate model chosen was Gaussian processes, as it combines robustness to overfitting and the well-documented accuracy of global kernel methods. It is a data-driven stochastic algorithm that models predictions as the posterior of the Bayes formula. The prior and the likelihood are modeled as Gaussian distributions whose parameters are optimized based on the already measured samples. In this way, the algorithm elegantly provides an inherent uncertainty quantification, which in data sets of small size can be crucial. A more detailed description can be found in SI or in ref. 63.
It must be noted that AUGUR aims at providing predictions and optimization with a limited dataset. If the complexity of the system rises significantly, one must resort to the so-called batch GP64 or fully Bayesian neural networks65. These algorithms should be able to handle large datasets, though at the cost of somewhat decreased performance. However, with state-of-the-art algorithms implemented in gpytorch66,67 Gps can easily handle datasets of a million samples with very limited approximations.
Graph Neural Networks
Gaussian processes are a versatile family of algorithms. Nevertheless, they suffer significantly with feature spaces of large dimensionality. Further, meaningfully representing a molecular structure using a set of scalar features is non-trivial. One data-driven algorithm, however, that provides a natural encoding for molecules is Graph Neural Networks (GNNs).
The inputs of this architecture are graph structures, which consist of nodes, edges, and edge attributes. Each node is a mathematical representation of an atom. Node features that are used in this work are the atomic radius, atomic mass, and electronegativity of each atom. Edges are tuples that describe which nodes are connected. Lastly, the edge attributes contain information about the strength of interaction between two nodes. In the present study, the relevant entry of the coulomb matrix and the pairwise distance between two atoms were used as edge features.
The resulting representations are processed with graph convolution layers. These layers operate in two steps. Initially, all the nodes connected to a specific node create messages that codify the effect of these nodes on the receiving one. The messages can be generated by a variety of algorithms, but in the present work, we chose a neural network inspired by ref. 68. This process is repeated for every node.
The second step is the aggregation process, where all the messages directed at one node are condensed into one update that is then applied to the node features. Typically, this can be done by averaging or selecting the maximum message values, but more involved strategies like transformers can be applied. The updated node features now contain information about their neighboring nodes, thus giving more context to the representation.
Finally, once sufficient context has been applied, the graph representation is pooled down to a fixed dimension vector. Typically, this can be done by averaging or selecting the maximum value of every feature across the entire graph. This step condenses a graph of arbitrary size down into a fixed number of representations. In this way, the same algorithm can process different molecules of various sizes without relying on suboptimal techniques.
Graph neural networks can naturally encode molecules but lose the stochastic nature of the Gaussian processes. To this end, we have used a GNN as a feature extractor from the molecules, and these features are used as inputs to the GP. The parameters of the full pipeline are trained simultaneously towards the goal of minimizing the marginal log likelihood. In this way, we combine the advantages of both algorithms.
Data availability
Data are available at https://github.com/benkour/AUGUR.
Code availability
Code are available at https://github.com/benkour/AUGUR.
References
Kouroudis, I., Gosswein, M. & Gagliardi, A. Utilizing data-driven optimization to automate the parametrization of kinetic Monte Carlo models. J. Phys. Chem. A 127, 5967–5978 (2023).
Lampe, C. et al. Rapid data-efficient optimization of perovskite nanocrystal syntheses through machine learning algorithm fusion. Adv. Mater. 35, 2208772 (2023).
Kollmannsberger, K. L. et al. Mechanistic insights into zif-8 encapsulation of atom-precise pt(m) carbonyl clusters. Chem. Mater. 35, 5475–5486 (2023).
Garlyyev, B. et al. Optimizing the size of platinum nanoparticles for enhanced mass activity in the electrochemical oxygen reduction reaction. Angew. Chem. Int. Ed. 58, 9596–9600 (2019).
Morris, G. M. et al. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Skúlason, E. et al. Density functional theory calculations for the hydrogen evolution reaction in an electrochemical double layer on the pt(111) electrode. Phys. Chem. Chem. Phys. 9, 3241–3250 (2007).
Wei, C. M., Groá, A. & Scheffler, M. Ab initio calculation of the potential energy surface for the dissociation of H2 on the sulfur-covered pd(100) surface. Phys. Rev. B 57, 15572–15584 (1998).
Jonsson, H., Mills, G. & Jacobsen, K. W. Nudged elastic band method for finding minimum energy paths of transitions. In Classical and quantum dynamics in condensed phase simulations, 385–404 (World Scientific, 1998).
Goedecker, S. Minima hopping: An efficient search method for the global minimum of the potential energy surface of complex molecular systems. J. Chem. Phys. 120, 9911–9917 (2004).
Schonborn, S. E., Goedecker, S., Roy, S. & Oganov, A. R. The performance of minima hopping and evolutionary algorithms for cluster structure prediction. J. Chem. Phys. 130, https://doi.org/10.1063/1.3097197 (2009).
Barducci, A., Bussi, G. & Parrinello, M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 100, 020603 (2008).
YazdanYar, A., Aschauer, U. & Bowen, P. Adsorption free energy of single amino acids at the rutile (110)/water interface studied by well-tempered metadynamics. J. Phys. Chem. C. 122, 11355–11363 (2018).
Martí, C. et al. Dockonsurf: A Python code for the high-throughput screening of flexible molecules adsorbed on surfaces. J. Chem. Inf. Model. 61, 3386–3396 (2021).
Pedretti, E., Restuccia, P. & Righi, M. C. Xsorb: A software for identifying the most stable adsorption configuration and energy of a molecule on a crystal surface. Comput. Phys. Commun. 291, 108827 (2023).
Mayr, F., Harth, M., Kouroudis, I., Rinderle, M. & Gagliardi, A. Machine learning and optoelectronic materials discovery: a growing synergy. J. Phys. Chem. Lett. 13, 1940–1951 (2022).
Lorenz, S., Scheffler, M. & Gross, A. Descriptions of surface chemical reactions using a neural network representation of the potential-energy surface. Phys. Rev. B − Condens. Matter Mater. Phys. 73, 115431 (2006).
Jager, M. O., Morooka, E. V., Federici Canova, F., Himanen, L. & Foster, A. S. Machine learning hydrogen adsorption on nanoclusters through structural descriptors. npj Computational Mater. 4, 37 (2018).
Pablo-García, S. et al. Fast evaluation of the adsorption energy of organic molecules on metals via graph neural networks. Nat. Comput. Sci. 3, 433–442 (2023).
Lan, J. et al. Adsorbml: a leap in efficiency for adsorption energy calculations using generalizable machine learning potentials. npj Comput. Mater. 9 https://doi.org/10.1038/s41524-023-01121-5 (2023).
Jinnouchi, R., Lahnsteiner, J., Karsai, F., Kresse, G. & Bok-dam, M. Phase transitions of hybrid perovskites simulated by machine-learning force fields trained on the fly with bayesian inference. Phys. Rev. Lett. 122, 225701 (2019).
Xie, Y. et al. Uncertainty-aware molecular dynamics from bayesian active learning for phase transformations and thermal transport in sic. npj Comput. Mater. 9, 36 (2023).
Wollschlager, T., Gao, N., Charpentier, B., Ketata M. A. & Gunnemann S. Uncertainty estimation for molecules: Desiderata and methods. In International Conference on Machine Learning, 37133–37156 (PMLR, 2023).
Yamashita, T. et al. Crystal structure prediction accelerated by bayesian optimization. Phys. Rev. Mater. 2, 013803 (2018).
Deshwal, A., Simon, C. M. & Doppa, J. R. Bayesian optimization of nanoporous materials. Mol. Syst. Des. Eng. 6, 1066–1086 (2021).
Zuo, Y. et al. Accelerating materials discovery with Bayesian optimization and graph deep learning. Mater. Today 51, 126–135 (2021).
Tran, K. & Ulissi, Z. W. Active learning across intermetallics to guide discovery of electrocatalysts for CO2 reduction and H2 evolution. Nat. Catal 1, 696–703 (2018).
Peterson, A. A. Acceleration of saddle-point searches with machine learning. J. Chem. Phys. 145 (2016).
Koistinen, O.-P., Ásgeirsson, V., Vehtari, A. & Jónsson, H. Nudged elastic band calculations accelerated with Gaussian process regression based on inverse interatomic distances. J. Chem. Theory Comput. 15, 6738–6751 (2019).
Jung, H., Sauerland, L., Stocker, S., Reuter, K. & Margraf, J. T. Machine-learning driven global optimization of surface adsorbate geometries. npj Comput. Mater. 9, 114 (2023).
Chan, L., Hutchison, G. R. & Morris, G. M. Bayesian optimization for conformer generation. J. Cheminform. 11 https://doi.org/10.1186/s13321-019-0354-7 (2019).
Todorovic, M., Gutmann, M. U., Corander, J. & Rinke, P. Bayesian inference of atomistic structure in functional materials. Npj Comput. Mater. 5, 35 (2019).
Järvi, J., Rinke, P. & Todorović, M. Detecting stable adsorbates of (1S)-camphor on Cu(111) with Bayesian optimization. Beilstein J. Nanotechnol. 11, 1577–1589 (2020).
Fang, L., Guo, X., Todorović, M., Rinke, P. & Chen, X. Exploring the conformers of an organic molecule on a metal cluster with Bayesian optimization. J. Chem. Inf. Model. 63, 745–752 (2023).
Raha, S. & Ahmaruzzaman, M. Zno nanostructured materials and their potential applications: progress, challenges and perspectives. Nanoscale Adv. 4, 1868–1925 (2022).
Dmytruk, A., Dmitruk, I., Shynkarenko, Y., Belosludov, R. & Kasuya, A. Zno nested shell magic clusters as tetrapod nuclei. RSC Adv. 7, 21933–21942 (2017).
Lei, D. et al. Ion-transport kinetics and interface stability augmentation of zinc anodes based on fluorinated covalent organic framework thin films. Adv. Energy Mater. 14, 2403030 (2024).
Cahangirov, S., Topsakal, M., Akturk, E., Cahin, H. & Ciraci, S. Two- and one-dimensional honeycomb structures of silicon and germanium. Phys. Rev. Lett. 102, 236804 (2009).
Takeda, K. & Shiraishi, K. Theoretical possibility of stage corrugation in Si and Ge analogs of graphite. Phys. Rev. B 50, 14916–14922 (1994).
Dmytruk, A. et al. Zno clusters: Laser ablation production and time-of-flight mass spectroscopic study. Microelectron. J. 40, 218–220 (2009).
Larsen, A. H. et al. The atomic simulation environment—a Python library for working with atoms. J. Phys.: Condens. Matter 29, 273002 (2017).
Daliran, S. et al. Defects and disorder in covalent organic frameworks for advanced applications. Adv. Funct. Mater. 34, 2312912 (2024).
Jimenez, M. P., Campos, J., Jover, J., Alvarez, S. & Carmona, E. Coordination of e–c bonds (e = zn, mg, al) and the zn–h bonds of (c5me5)znh and (c5me5)znznh across a quadruply bonded dimolybdenum dihydride complex. Organometallics 41, 3225–3236 (2022).
Bosi, F., Andreozzi, G. B., Halenius, U. & Skogby, H. Zn-o tetrahedral bond length variations in normal spinel oxides. Am. Mineral. 96, 594–598 (2011).
Dong, H. et al. Two-coordinate NHc-supported ZnII organocations: Steric and electronic tunability and use in alkyne hydroboration catalysis. Eur. J. Inorg. Chem. 2022, e202101002 (2022).
Huang, Z. et al. Structures, properties and application of 2d monoelemental materials (xenes) as graphene analogues under defect engineering. Nano Today 35, 100906 (2020).
Gao, J., Zhang, J., Liu, H., Zhang, Q. & Zhao, J. Structures, mobilities, electronic and magnetic properties of point defects in silicene. Nanoscale 5, 9785–9792 (2013).
Sivek, J., Sahin, H., Partoens, B. & Peeters, F. M. Adsorption and absorption of boron, nitrogen, aluminum, and phosphorus on silicene: Stability and electronic and phonon properties. Phys. Rev. B 87, 085444 (2013).
Pham, H. D., Gumbs, G., Su, W.-P., Thuy Tran, N. T. & Lin, M.-F. Unusual features of nitrogen substitutions in silicene. RSC Adv. 10, 32193–32201 (2020).
Hernández Cocoletzi, H. & Castellanos Águila, J. Dft studies on the al, b, and p doping of silicene. Superlattices Microstruct. 114, 242–250 (2018).
Das, R., Chowdhury, S., Majumdar, A. & Jana, D. Optical properties of p and al doped silicene: a first principles study. RSC Adv. 5, 41–50 (2015).
Wang, X., Liu, H. & Tu, S.-T. Study of formaldehyde adsorption on silicene with point defects by DFT method. RSC Adv. 5, 65255–65263 (2015).
White, E. W. & Brown, J. M. Variation of mean Si-O bond lengths in silicon-oxygen octahedra. J. Solid State Chem. 24, 117–125 (1977).
Bragg, R. H. & Corbett, M. C. Silicon-oxygen bond lengths, bridging angles si-o-si and synthetic low tridymite. Acta Crystallogr. Sect. B: Struct. Crystallogr. Cryst. Chem. 33, 1234–1242 (1977).
Marshall, F. J. & Gibson, K. F. A computational characterization of boron-oxygen multiple bonding in Hn=Ch-Ch=Ch-Nh-Bo. J. Phys. Chem. A 112, 3467–3476 (2008).
Cruickshank, F. The b-o bond lengths in boron-oxygen compounds. Acta Crystallogr. Sect. B: Struct. Crystallogr. Cryst. Chem. 24, 228–235 (1968).
Basolo, F. & Pearson, R. G. Carbon–oxygen bond energies and bond distances. J. Phys. Chem. 61, 606–609 (1957).
Weller, M. T. & Jones, R. G. Bond length-bond valence relationships for carbon-carbon and carbon-oxygen bonds. J. Adv. Anal. Sci. 12, 189–197 (2015).
Mulliken, R. S. & Person, W. B. Molecular parameters and bond structure—v. nitrogen—oxygen bonds. Spectrochim. Acta Part A: Mol. Spectrosc. 22, 753–773 (1966).
Waters, J. M. & Gray, H. B. Lengths of phosphorus-oxygen and sulfur-oxygen bonds. An extended Hückel molecular orbital examination of Cruickshank’s dr-pt. Inorg. Chem. 7, 638–645 (1968).
Demidov, P. V. & Pudovik, A. N. Distortion of opo bond angles in phosphorus monoanions: Ab initio studies. J. Inorg. Biochem. 59, 93–102 (1995).
Ruiz, J., McAdon, M. & Garcés, J. Aluminum complexes as models for brønsted acid sites in zeolites: Structure and energetics of [Al(oh)4]-, [Al(H2O)6]3+, and intermediate monomeric species [Al(Oh)x(H2O)n-x⋅mH2O] 3-x obtained by hydrolysis. J. Phys. Chem. B 101, 1733–1744 (1997).
Kelber, J. A. & Cabrera, C. R. Probing the atomic-scale structure of amorphous aluminum oxide grownby atomic layer deposition. ACS Appl. Mater. Interfaces 12, 1672–1684 (2020).
Williams, C. K. I. & Rasmussen, C. E. Gaussian Processes for Machine Learning, 2 (MIT Press Cambridge, MA, 2006).
Daxberger, E. A. & Low, B. K. H. Distributed batch Gaussian process optimization. In International Conference on Machine Learning, 951–960 (PMLR, 2017).
Kononenko, I. Bayesian neural networks. Biol. Cybern. 61, 361–370 (1989).
Gardner, J., Pleiss, G., Weinberger, K. Q., Bindel, D. & Wilson, A. G. Gpytorch: Blackbox matrix-matrix gaussian process inference with gpu acceleration. Adv. Neural Inf. Process. Syst. 31, 7576–7586 (2018).
Pleiss, G., Gardner, J., Weinberger, K. & Wilson, A. G. Constant-time predictive distributions for Gaussian processes. In International Conference on Machine Learning, 4114–4123 (PMLR, 2018).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In International Conference on Machine Learning, 1263–1272 (PMLR, 2017).
Acknowledgements
The authors acknowledge funding from the project ProperPhotoMile, supported under the umbrella of SOLAR–ERA.NET Cofund 2 by The Spanish Ministry of Science and Education and the AEI under the project PCI2020-112185 and CDTI project number IDI–20210171; the Federal Ministry for Economic Affairs and Energy on the basis of a decision by the German Bundestag project number FKZ 03EE1070B and FKZ 03EE1070A; and the Israel Ministry of Energy with project number 220-11-031. SOLAR-ERA.NET is supported by the European Commission within the EU Framework Program for Research and Innovation HORIZON 2020 (Cofund ERA-NET Action, 786483). Further, A.G. acknowledges financial support from TUM Innovation Network for Artificial Intelligence powered Multifunctional Material Design (ARTEMIS) and funding in the framework of Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy - EXC 2089/1 - 390776260 (e-conversion). Lastly, we wish to express our gratitude to Dr. Inigo Iribarren for creating the flowcharts for this work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
I.K.: Conceptualization, Formal Analysis, Data Curation, Validation, Investigation, Methodology, Project Administration, Software, Supervision, Writing P: Conceptualization, Data Curation, Validation, Formal Analysis, Software Investigation, Methodology, Visualization, Writing N.M.: Data Curation, Formal Analysis, Investigation, Software, Methodology FM: Software, Methodology, Validation, Visualization, Writing (original draft) L.M.: Data Curation, Formal Analysis, Investigation, Methodology, Writing Z.G.: Data Curation, Formal Analysis, Software, Investigation A.G.: Resources, Funding, Supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kouroudis, I., Poonam, Misciasci, N. et al. AUGUR, a flexible and efficient optimization algorithm for identification of optimal adsorption sites. npj Comput Mater 11, 136 (2025). https://doi.org/10.1038/s41524-025-01630-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41524-025-01630-5