Abstract
Automating structured data extraction from scientific literature is a critical challenge with broad implications across domains. We introduce nanoMINER, a multi-agent system combining large language models and multimodal analysis to extract essential information from scientific research articles on nanomaterials. This system processes documents end-to-end, utilizing tools such as YOLO for visual data extraction and GPT-4o for linking textual and visual information. At its core, the ReAct agent orchestrates specialized agents to ensure comprehensive data extraction. We demonstrate the efficacy of the system by automating the assembly of nanomaterial and nanozyme datasets previously manually curated by domain experts. NanoMINER achieves high precision in extracting nanomaterial properties like chemical formulas, crystal systems, and surface characteristics. For nanozymes, we obtain near-perfect precision (0.98) for kinetic parameters and essential features such as Cmin and Cmax. To benchmark the system performance, we also compare nanoMINER to several baseline LLMs, including the most recent multimodal GPT-4.1, and show consistently higher extraction precision and recall. Our approach is extensible to other domains of materials science and fields like biomedicine, advancing data-driven research methodologies and automated knowledge extraction.
Similar content being viewed by others
Introduction
The exponential growth of scientific literature in recent decades has created an unprecedented challenge: efficiently extracting, structuring, and making accessible the vast scientific knowledge concentrated in research papers. The challenge is especially significant in fast-growing fields like materials science, where discoveries outpace manual data processing. Automated systems for data extraction have become essential.
Recent breakthroughs in natural language processing (NLP), particularly in Large Language Models (LLMs), have shown promise in addressing this challenge. These developments dramatically improved the precision of named entity recognition (NER) and relation extraction (RE)1. The emergence of self-attention mechanisms and LLMs has significantly enhanced natural language understanding and generation capabilities2,3,4,5,6,7,8. Among the notable developments in this field are models like Mistral-7B9, Llama-3-8B10, and GPT11, which are trained on large datasets for various NLP tasks. More recently, the introduction of multi-modal models like GPT-4V12,13 and GPT-4 Omni14,15 has extended these capabilities to include processing image and audio alongside text, opening new avenues for multi-model data processing. The potential of these advanced models is particularly promising in material science, where the complexity of chemical nomenclature and the prevalence of cross-domain terminology present unique challenges for automated data extraction16. As materials science embraces data-driven discovery methods, LLMs are perceived as indispensable tools for automating the extraction of critical experimental details, including chemical formulas and material properties, from unstructured text1. Recent benchmarks like SciDaSynth17 have demonstrated significant progress in structured knowledge extraction from scientific literature, leveraging large multimodal models (LMMs) to process data interactively and distill information from text, tables, and figures into structured formats. Similar success has been observed in clinical data, where NLP frameworks integrated with advanced LLMs have achieved accuracy in extracting complex data from electronic health records while significantly improving efficiency over traditional manual review methods18.
Despite the demonstrated capabilities of LLMs in chemistry and materials science, their performance is still not flawless19,20,21. The language used in the corresponding research articles is rather specific, combining chemical formulas, physical and chemical characteristics of substances, and their biological activity. In creating the CHEMDNER resource21, the authors specialize in chemical entities, such as processing signs, specialization, abbreviations, first-seen substances, and establishing boundaries. Another notable approach in this field is the NERRE method, which focuses on extracting chemical terms from text22. Such approaches focus on processing only small text segments, potentially missing the broader context in full documents. Moreover, similar systems require significant human intervention, particularly for interpreting figures and supplementary materials, limiting the extent of automation. In nanozymes, for example, a study by Sun et al.23 explores ChatGPT alongside traditional machine learning to support the extraction and prediction of nanozyme catalytic activities. They achieved a 65.11% similarity between ChatGPT-extracted data and manually curated datasets, demonstrating the potential of LLMs for processing text-based information from the scientific literature. However, their approach cannot differentiate between individual experiments within a single article and requires human experts to interpret figures and supplementary materials. This further highlights the need to refine existing approaches to fully automated, high-precision extraction.
It is also worth mentioning that in the most recent study, Lee et al.24 used a pre-trained LLaMa-2 model with NER and RE to extract synthesis parameters of gold nanoparticles, significantly reducing the reliance on manual data collection. There, a standardized database with 492 formulations was established, and the model was able to predict the crystal system of nanoparticles based solely on the chemical formula, indicating the ability of LLaMa to perform zero-shot classification of materials. A similar approach utilizing GPT-4 was described in the work by Ansari and Moosavi25 that introduced the Eunomia agent, enabling the extraction of information on MOF materials and their properties using an LLM without the need for prior training. Despite these advances, automated extraction of scientific knowledge from research literature still faces several critical challenges. Current approaches demonstrate limited capability in processing complete research articles, struggle with integrating multiple data modalities, and require substantial human intervention to maintain high precision17,23,24. Particularly challenging is the automated processing of comprehensive experimental contexts, including figures and supplementary materials, while differentiating between individual experiments within the same article. These limitations significantly impede the scalability and efficiency of automated data extraction systems in materials science.
To address these fundamental challenges, we introduce nanoMINER, a multi-agent system designed to automate structured data extraction from scientific literature. Our system integrates state-of-the-art LLMs with specialized analysis tools to process entire complex research articles, maintaining full context and reducing the need for human intervention. At the core of our system is the multi-agent architecture centered around the main agent, which orchestrates various specialized agents for different aspects of data extraction, i.e., vision and NER agents. While simpler pipeline setups could potentially accomplish tasks like text or graph processing independently, our multi-agent system offers advantages in modularity, parallelization, and error handling. In particular, having a dedicated coordinator agent enables more flexible interactions between vision and text processing, especially when reconciling figure-derived data with textual descriptions. Notably, our system design is in agreement with the recent findings26 in multi-agent orchestration, showing that a coordinating agent is essential for multi-task processing pipelines as it improves overall performance and supports system extensibility.
The capabilities of the proposed system are demonstrated on two datasets of nanomaterials characterized by 7 and 10 parameters, respectively. The first dataset demonstrated the system’s ability to extract common nanomaterial characteristics, such as formula, size, surface modification, and crystal system. The second dataset focused on specific experimental properties characterizing enzyme-like activity.
NanoMINER integrates natural language processing, computer vision, and named entity recognition, achieving precision no less than 0.96 for Km, Vmax, minimal and maximal substrate concentrations in the nanozyme data. For nanomaterials, it demonstrated up to 0.66 precision in coating molecule weight extraction and close-to-zero normalized Levenshtein distances for the chemical formulas and coating molecules. Interestingly, nanoMINER was able to infer crystal systems from chemical formulas, showcasing its ability to extract explicit and implicit data and deliver data-driven insights from complex scientific data. Finally, we evaluate nanoMINER by comparing it to the top-rated LLMs as strong baselines, including the latest multimodal GPT-4.1 and the two reasoning models, namely, o3-mini and o4-mini. Despite using an older model for the Main agent (GPT-4o), our approach consistently outperformed the baselines in average precision, recall and F1 score. This highlights the benefits of decomposing automated information extraction into several smaller tasks delegated to coordinated, fine-tuned agents, and underscores the importance of agent orchestration. Our results hold promise for the future of accelerated research in natural sciences due to streamlined processes of data gathering and curation.
Results
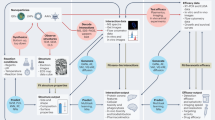
Automated structured data extraction from unstructured scientific literature is a challenging yet pivotal task with far-reaching impacts across scientific domains. We’ve developed nanoMINER, a multi-agent system for extracting structured data from scientific literature, focusing on nanomaterials. The system includes specialized agents for processing text and visual information from research papers and supplementary materials. The main agent coordinates the workflow, initiating text data extraction and interacting with other agents based on the content type. In this process, text data extraction refers to the Main agent retrieving and analyzing the full text of the article before receiving inputs from the NER and Vision agents, as shown in Fig. 1. This ensures that the complete article content is considered before structuring the extracted information.
The NER agent extracts key entities from text, while the vision agent analyzes visual data. The system integrates outputs from both agents to generate structured data, including material compositions, surface modifiers, reaction conditions, and catalytic properties, which are then summarized for straightforward interpretation and further analysis.
Main agent aggregates information obtained from NER and Vision agents. At the final stage of the pipeline, the aggregated information is processed to ensure a structured and consistent output format.
Automated extraction pipeline
Our system starts by processing input PDF documents containing nanomaterials literature (Fig. 1). Specialized tools extract text, images, and plots from the PDFs, ensuring all data modalities are captured (see PDF processing of the Methods). We employ the YOLO model27 for visual data extraction, specifically designed to detect and identify objects within images, such as figures, tables, and schemes. The extracted visual information is then analyzed by GPT-4o14,15, linking it with textual descriptions for a cohesive interpretation. After initializing the tools available to the main agent, the textual content undergoes strategic segmentation into 2048 chunks to facilitate efficient processing by different system components. The tool set includes named entity recognition using fine-tuned Mistral-7B and Llama-3-8B modes, figure processing with GPT-4o and YOLO, and full-text analysis with GPT-4o by the main agent. The original article text is split into segments of 2048 tokens to extract named parameters optimized for the overall extraction quality. The core of our system is the ReAct28 agent based on GPT-4o, which manages the other agents, performs function-calling, and merges the incoming information.
We developed an NER agent based on an LLM, which we further trained to extract essential classes from articles on nanomaterials. The system’s performance improved in tandem with the main agent, benefiting from feedback provided by the NER agent. We experimented with models such as Mistral-7B9 and Llama-3-8B10, whose sizes were chosen based on their ease of integration into a holistic system29. Agent based on Mistral-7B is designed to extract critical parameters for specialists. The generated dictionary allows the main agent to focus on the values highlighted by this agent, ensuring high precision across the entire agent system.
Based on GPT-4o, the vision agent enables precise processing of graphical images and non-standard tables in the articles, which standard PDF text extraction tools cannot parse. Our experiments have shown that some information and parameter values are only presented in charts, and this tool provides the main agent with additional information in such cases, improving the extraction quality. We tested nanoMINER on two datasets and presented the results in the following sections.
Nanomaterials data extraction
First, our system was evaluated on a subset of the nanomaterials dataset from the DiZyme30 database. It comprises 19 articles with 25 unique experiments, each manually annotated with key nanomaterial parameters such as formula, crystalline system, sizes, and surface modification. Extraction performance was tested using precision and recall metrics for the numerical parameters (Table S1) and the normalized Levenshtein distance for the categorical parameters, as detailed in Methods. Two configurations were tested for each parameter: (i) text-only extraction using GPT-4o, (ii) text extraction using GPT-4o, and NER using Mistral and Llama together to leverage their collective intelligence. Although the combined ‘Text+NER’ approach typically yields modest improvements in precision (e.g., for Mw(coating), precision increases from 0.62 to 0.66), it provides more pronounced gains in recall (e.g., Mw(coating) from 0.73 to 0.86). This indicates that the NER-based pipeline captures sparse or implicit references more effectively, thereby enhancing the overall robustness of the extraction process. There was no need to include a vision agent in nanomaterials, as all relevant data is typically found in text. Each configuration was tested 100 times to assess reproducibility, with slight variations in results stemming from the system’s interpretation of text and figures during each run. Figure 2 presents the average scores, with detailed precision metrics (mean ± standard deviation) available in Table S1.
The performance of numerical nanomaterial parameters extraction from scientific papers using different multi-agent system configurations (Text only and Text+NER) was calculated based on 25 experiments from 19 test papers. Average precision (A) and recall (B) of the numerical nanomaterial parameter extraction are presented. Performance of the main agent in сategorical nanomaterial parameter extraction using normalized Levenshtein distance is depicted for formula (C), crystal system (D), and surface (E) parameters.
Good precision (0.62–0.66) and the highest recall (0.73–0.86) across all configurations were demonstrated for the molecular weight coating parameter (“Mw(coat)”). The length, width, and depth parameterы also showed relatively good extraction performance, with precision (0.89–0.90) and recall (0.60–0.62). The typical reporting practices of nanoparticle morphology drive these empirical results. For spherical nanoparticles, papers frequently report a single “size” or “diameter” rather than units of three orthogonal dimensions. By default, nanoMINER tends to ignore cases when there is ambiguity or incomplete information regarding the dimensions of the nanoparticle morphology, thus lowering recall values.
Using the normalized Levenshtein distance, we evaluated the extraction accuracy of string parameters—Formula, Crystal system, and Surface parameters. The Formula parameter (Fig. 2C) performed best, with distances centered around zero. The Surface parameter (Fig. 2E) showed similar performance but had a heavy tail in the distance distribution due to variations in molecular name formats. The crystal system parameter (Fig. 2D) displayed a bimodal distribution, indicating instances differentiated from human annotations, while the system’s accurate predictions contributed to the higher distances. Initially, we hypothesized that the main agent used JCPDS numbers to infer the crystal system; however, further investigation revealed that the crystal system was derived directly from chemical formulas. Additional tests on 100 randomly selected experiments yielded about 86% accuracy in predicting crystal systems solely based on chemical formulas.
In addition, we evaluated the system’s overall performance using the Jaccard index on a dataset of 656 nanomaterials experiments (Table S2) based on nanomaterials sets from the DiZyme database. This dataset specifically focuses on general nanomaterial parameters relevant to various downstream applications related to enzymatic, antibacterial, cytotoxic, and magnetic properties.
The Jaccard index measures similarity between sets with varying element counts. In the recent study by Chataut et al.31, the index was used to assess keyword extraction accuracy, where GPT-3.5 achieved scores of 0.64 on the Inspec dataset and 0.21 on the PubMed dataset. Our results showed that most parameters achieved Jaccard index values above 0.50, indicating strong alignment between manually and automatically extracted data. The Jaccard index provides a stricter measure than precision or recall, penalizing minimal discrepancies between extracted and actual parameters. Nevertheless, our system achieved scores of 0.54 for the Formula and Crystal systems and 0.69 for the Surface. Combined with the precision, recall, and Levenshtein distance presented above, these results underscore nanoMINER’s ability to handle complex experimental data from nanomaterials literature accurately.
Nanozymes data extraction
Building upon nanomaterial data extraction, we expanded our multi-agent system to the specialized field of nanozymes, a new class of nanomaterials with enzyme-like properties. This transition highlights the adaptability of our system, bridging fundamental material properties with advanced applications in bionanotechnology. Nanozymes combine nanotechnology and enzymology, exhibiting catalytic activities similar to natural enzymes. To thoroughly capture the characteristics of these artificial enzymes, we identified and extracted ten critical parameters, including catalytic activity type, kinetic constants, and reaction conditions. These parameters create a comprehensive profile of nanozyme performance, enabling researchers to compare different nanozymes, optimize reaction conditions, and understand structure-function relationships. We tested three configurations in our experiments: (i) text-only extraction, (ii) text and vision processing, and (iii) text, vision, and NER. Although ‘Text+Vision’ already significantly boosts precision for parameters typically found in graphs (such as C min and C max, where precision rises from 0.90 to 0.97 and 0.91 to 0.98, respectively), adding NER in ‘Text+Vision+NER’ sometimes yields only marginal additional gains in precision (e.g., from 0.68 to 0.70 for Temperature) or no improvement at all. This is because the Vision agent already captures most of the graph-based data that drives precision upward, and the textual references to those same parameters are usually consistent enough that NER does not further reduce false positives. However, NER typically provides a stronger boost to recall (e.g., for Km from 0.88 to 0.91 and Temperature from 0.88 to 0.96), as it helps identify less obvious mentions or synonyms across the text. Thus, the value of NER lies in retrieving sparse or context-dependent data rather than substantially raising precision for parameters already well-detected in graphs. Including the Vision agent proved crucial for extracting specific properties, such as concentration parameters, often represented only in figures. In the case of nanozymes, this combined approach was convenient in retrieving data that was otherwise absent from the main text but visually depicted on graphs or charts. Our system demonstrated remarkable proficiency in extracting critical enzymatic parameters across 56 experiments with peroxidase, oxidase, and catalase activities. Each configuration was tested 100 times to account for slight variations in the results between runs. Values were averaged across these experiments, and the standard deviation (std) was calculated to assess variability (Table S3). NanoMINER achieved excellent precision for the majority of parameters, with high scores ranging from 0.90 to 0.98 for C min and C max. Extraction of Km, Vmax, and pH also exhibited high precision, with values of 0.97 for Km, 0.96 for Vmax, and 0.89 for pH across configurations (Fig. 3). Temperature extraction yielded the lowest precision among all parameters, reaching only 0.68. Recall scores were generally lower than precision, as commonly observed in nanomaterial parameter extraction experiments. However, temperature recall was the highest among all parameters, reaching up to 0.96. These precision and recall values for temperature can be explained by the fact that temperature is not described in many experiments, and the system sometimes attempts to substitute room temperature, which we counted as an error. High recall scores were also observed for Km, ranging from 0.87 to 0.91, and Vmax, ranging from 0.79 to 0.83, across different configurations. Recall for concentration parameters (C min, C max, C co-sub) was relatively lower, ranging from 0.38 to 0.54, likely because these parameters frequently appear on figure axes but are not always mentioned in the main text of the papers. Similar to our findings in nanomaterials extraction, the Text+Vision+NER configuration generally outperformed the other configurations, particularly in recall. This underscores the value of integrating visual information processing and named entity recognition in scientific text analysis. We utilized normalized Levenshtein distance to evaluate the extraction of categorical nanozyme parameters. Both catalytic activity (Fig. 3C) and reaction type (Fig. 3D) parameters exhibited an unimodal distribution centered around zero, indicating consistently reliable extraction performance. The long tails of the distributions reaching the Levenshtein distance of one were attributed to rare partial mismatches between the extracted text and the reference parameters.
The performance of numerical nanozyme parameter extraction from scientific papers using different multi-agent system configurations (Text only, Text+Vision, and Text+Vision+NER) was calculated based on 56 experiments from 19 test papers. Average precision (A) and recall (B) of nanozyme data extraction are presented. The performance of сategorical nanozyme parameter extraction using normalized Levenshtein distance is depicted for catalytic activity (C) and reaction type (D) parameters.
These errors often arose from slight variations in terminology that nanoMINER needed help reconciling fully. For instance, synonyms or alternate phrasing for catalytic activity and reaction types occasionally led to minor discrepancies, resulting in a Levenshtein distance of one rather than a perfect match.
To ensure robust evaluation, we additionally measured the Jaccard index on a complete dataset of 1177 nanozyme experiments (Table S4) coming from the DiZyme30 database. This metric assesses how well the system replicates the structure and complexity of real-world data, providing a more holistic view of performance. Strikingly, we achieved the Jaccard index score of 0.77 for pH, representing the highest level of consistency with manually curated data across all parameters. Additionally, the score of 0.70 for Activity further indicates high overall consistency, while the score of 0.61 for Reaction type reflects the system’s ability to capture specific experimental details reliably. Our empirical results suggest that nanoMINER can handle intricate scientific data extraction tasks effectively. Given the complexity of the nanozyme domain, these findings look impressive and promising for the future of automated knowledge extraction systems.
Comparison to baselines
To evaluate the benefits of our multi-agent system over simpler approaches, we compared nanoMINER against several strong baselines: the latest multimodal GPT-4.1, as well as two reasoning models o3-mini and o4-mini. These models were applied directly to the full text extracted from the PDF with no additional processing steps. We used the same prompts and instructions as those in nanoMINER. All parameters were extracted with a single pass.
NanoMINER achieved the highest average performance metrics (Table 1), despite using an older GPT-4o model with no reasoning for the Main agent. This highlights the advantage of decomposition of the complex information extraction task into visual data analysis and named entity recognition handled by individual agents.
Notably, the only parameters for which the advantage of nanoMINER is less pronounced are geometric dimensions, i.e., Length, Width, and Depth (Table S5). This is due to domain-specific reporting practices: many nanomaterials are spherical, and authors often report a single diameter value without distinguishing between axes. As a result, extracting separate dimensional parameters may lead to ambiguity or over-segmentation in cases where a single size descriptor is intended. These challenges do not reflect system limitations per se but rather inconsistencies in source reporting that can be explicitly accounted for in the future.
While the baseline models provided strong LLM-based extraction capabilities, we observed substantial variation in their output formatting. GPT-4.1 produced structured responses similar to GPT-4o, but often appended auto-generated tables or summaries at the end. By contrast, o3-mini and o4-mini generated outputs in less standardized formats, with experiment descriptions embedded in long narrative sections. This made it significantly harder to identify discrete experimental units or link parameter values correctly. For example, concentrations, reaction tracks, and kinetic constants were often nested within free-text descriptions like “all assays were carried out in acetate buffer…” or only referenced through phrases such as “see Table 1 of the article”. This inconsistency highlights the practical challenges of using foundation models without orchestration or model-specific prompt engineering for scientific extraction tasks. In contrast, nanoMINER processes the entire article context with coordination between specialized agents, allowing for better segmentation of experiments, association of parameters, and interpretation of non-standard structures like embedded figures and captions.
Validation case study
To illustrate the system’s capabilities overall, we selected a paper with four individual experiments32 as a validation case. We manually collected the target parameters from the original text and compared them to those extracted by our multi-agent system. A complete comparison is given in Table 2. The results demonstrate high concordance between manual and automated extraction across parameters. Notably, our system achieved this level of precision in under a minute (excluding image processing time), compared to approximately 90 minutes for manual extraction by lab experts. The results in Table 2 highlight critical parameters such as Formula, Activity, Km, and Vmax, accurately extracted and matching the manually collected values. For example, the system correctly identified the chemical formula “Ru”, reported particle sizes (“20–30 nm”), and catalytic activities like “POD” (peroxidase) and “OXD” (oxidase) for the respective experiments.
We found a discrepancy in the Ccat (mg/mL), where the automated extraction yielded “10” in some experiments while the actual value was “50”. Upon closer examination, this difference is attributed to the same parameter measured for other assays, indicating that nanoMINER is sensitive to context and may capture matching data from other paper sections. Notably, parameters not explicitly stated in the paper, such as Crystal system, pH, and Temp, were consistently reported as “None” by manual and automated methods, reflecting the system’s ability to handle missing information appropriately.
This validation case underscores nanoMINER’s capability to accurately extract detailed experimental data from new, unseen documents, highlighting its generalization power and applicability. The high concordance with manual annotations across multiple parameters demonstrates the system’s proficiency in handling complex scientific texts and extracting nuanced information.
Discussion
Our research presents a practical contribution to the automated extraction of structured data from scientific literature, particularly in nanomaterials science. We have developed nanoMINER, a multi-agent system that leverages large language models, multimodal analysis, and retrieval-augmented generation to extract relevant information from research articles. The effectiveness of nanoMINER was demonstrated through testing on two representative datasets and a validation case study. The system’s performance was evaluated using precision, recall, Jaccard index, and the normalized Levenshtein distance to capture both numerical and categorical extracted parameters. The system achieved up to 0.98 precision and 0.96 recall across numerical parameters and accurate extraction of categorical data, as shown by the unimodal near-zero distribution of Levenshtein distances.
Built-in multimodality of the latest models holds the promise of a single LMM capable of solving automated data extraction from texts, tables, graphs, videos, etc. However, the performance of GPT-4o, as one of the most powerful multimodal models to date, in graph data extraction largely varies with visualization complexity. While the model excels at processing standard graphs with clear data presentation, it faces challenges with complex multi-series plots and non-standard formats. We provide detailed examples of both successful and challenging cases in the SI Graph Data Extraction Performance Section. Our empirical results employing other state-of-the-art multimodal and reasoning LLMs also confirmed the necessity of a modular, targeted approach to automated information extraction. This not only sheds light on limitations of the existing AI solutions, but also highlights the actuality of further development of agentic approaches.
During evaluation, we observed that nanoMINER demonstrated a notable capability to associate chemical formulas with their corresponding crystal systems, achieving 86% accuracy in these predictions. While human annotators typically marked the crystal system as missing when not explicitly reported, nanoMINER showed consistent performance in inferring this information from chemical formulas. This capability, while significant, likely reflects the language models’ training on scientific literature, where such associations are commonly present. The high accuracy and reproducibility of these predictions suggest this is more than mere hallucination, warranting further investigation. However, we acknowledge the need for additional validation across a comprehensive range of chemical formulas and crystal systems to fully characterize this capability. Future work will include gathering additional validation datasets and exploring similar off-target inference tasks to better understand this behavior in a broader context. This finding underscores both the potential and the need for careful validation of emergent capabilities in automated information extraction systems.
Being a fully automated information extraction tool, nanoMINER represents a leap forward from manual data collection. However, its performance remains imperfect. One limitation is the lack of consistency in extraction results for some parameters, indicated by high standard deviations in recall, e.g., for extracted concentrations of nanozymes. This is likely due to the diverse presentation of concentrations, often embedded in figures or supplementary materials rather than in the main text. Those inconsistencies can be mitigated by averaging repeated extraction experiments, prompt engineering, and reconfiguring the system to better handle particularly complex extraction tasks. Ultimately, randomized manual data checks seem absolutely necessary to ensure the reliability of the extraction. In cases of low performance metrics, follow-up data curation is strongly recommended. While the achieved precision values (up to 0.66 for certain parameters) may not reach near-perfect levels, they are sufficient for many downstream tasks such as automated literature screening, initial database population, and hypothesis generation. For more critical or safety-sensitive applications, we recommend incorporating a human-in-the-loop approach where extracted data undergoes selective expert review, particularly when coverage or complexity is high. Current levels of precision serve as a practical starting point for large-scale data extraction, with iterative improvements and domain-specific post-processing workflows further raising confidence in the final dataset.
Adaptability of our multi-agent architecture shows a potential for generalization to other scientific subdomains. By extending nanoMINER to retrieve magnetic, antibacterial, therapeutic, and other properties of nanomaterials, we envision the creation of the comprehensive nanomaterial database that can accelerate future scientific discoveries and industrial applications.
To achieve that ambitious goal, a deeper exploration of optimal system architecture might be required. With the emergence of highly capable open-source large language models, we consider fine-tuning a viable alternative to retrieval-augmented generation with market-leading models, offering on-par performance and reduced costs. Moreover, the future of automated information extraction will likely be a competition between multi-agent systems and individual multimodal models. Therefore, we keep an open mind to the design of the next generation nanoMINER.
Despite current limitations, nanoMINER effectively addresses nanomaterial information extraction from scientific articles. This work provides a step towards more efficient, data-driven research in materials science.
Methods
Data preprocessing
The procedure of annotation of articles for name entity recognition took place on the doccano platform33. Twelve articles about nanoparticles with peroxidase activity were selected for training, and a complete set of data was needed to fill a table in the database, based on which DiZyme service works. The team included chemists and materials scientists. Before starting, the practitioners were trained, and a guideline for article annotation was drawn up.
Nineteen papers with the most features included were annotated for the test set. Each example inside the paper was annotated separately. We selected these specific sizes (12 for training and 19 for evaluation) because they represent a manageable yet diverse set of articles in the nanomaterials domain, ensuring coverage of the parameter variability we target.
PDF processing
Pdfplumber34 was used to extract text and character information from PDF documents, focusing on maintaining the document’s layout and formatting. When pdfplumber encounters pages with insufficient text extraction due to embedded images or scanned content, py-tesseract performs optical character recognition (OCR) on the page images, ensuring no textual information is lost. The system also includes custom functions to handle footnotes, references, and character-to-symbol mapping, enhancing the overall precision and readability of the extracted text.
-
PDF Parsing with pdfplumber: Extracts structured text and character information, preserving the layout and formatting.
-
OCR with pytesseract: Performs text recognition on page images where direct text extraction is ineffective.
-
Text and Symbol Mapping: Ensures accurate character-to-symbol mapping to maintain text integrity.
-
Footnote and Reference Filtering: Excludes footnotes and references based on font size and name criteria.
LLM extraction
To ensure robustness and efficient processing, we segmented the articles into fragments of 2048 tokens, allowing us to feed the extracted text to the main agent later. Before dividing the article, we relied on the annotation to determine which classes were present in each chunk. The values of these classes were set as the target result for output, while the remaining nanozyme parameters in that fragment were considered empty. In this way, we augment the model’s dataset and achieve better generalization capability during inference, as we will only sometimes strive to find a value for every class.
The use of LLMs in information extraction tasks has gained significant attention in recent years35,36. Transformer-based models, such as BERT37 and GPT, have demonstrated remarkable performance in understanding and generating human-like text. By leveraging the power of these models and adapting them to the specific domain of nanomaterials, we aim to automate the process of extracting relevant information from scientific articles, facilitating faster and more efficient knowledge discovery in this field. In this study, we propose a multi-agent system designed to extract and structure information from chemical articles on nanozymes.
The main agent is equipped with the following tools:
Full-text Analysis. For the core text analysis and orchestration, we utilized GPT-4o due to its advanced capabilities in understanding complex scientific content and managing multi-agent interactions. This model’s ability to process both textual and visual information made it particularly suitable for our system’s needs. Named Entity Recognition. We utilized fine-tuned Mistral-7B and Llama-3-8B models. For text processing and named entity recognition, we chose Mistral-7B and Llama-3-8B; these models offered strong performance on our domain-specific corpus—comparable to larger (13B+) LLMs—while being significantly lighter in terms of memory and compute requirements. This aligns with studies in low-resource NER4,6, showing that mid-sized language models can effectively handle specialized tasks when properly fine-tuned. Graph Processing. We employed GPT-4o and YOLO for analysis of graphical elements. We implemented YOLOv8 for figure detection due to its robust object-detection accuracy27. Its single-pass architecture and proven performance on a wide range of images—including scientific documents—make it a suitable choice for extracting figures, tables, and schemes from complex PDF layouts.
Main agent
The core of our multi-agent system is an assistant based on GPT-4o, which extracts data from the article text. The full text of the prompt: “You are a helpful assistant in chemistry, specializing in nanozymes. Your task is to analyze scientific articles and extract detailed information about various experiments with nanozymes. It is crucial for you to accurately and comprehensively describe each experiment separately, without referring to other experiments in the article. Usually, the articles contain several experiments with nanozymes with different parameters, such as formula, activity (usually peroxidase, oxidase, catalase, or laccase), crystal system (usually cubic, hexagonal, tetragonal, monoclinic, orthorhombic, trigonal, amorphous, or triclinic), length, width, and depth (or just size or diameter), surface chemistry (naked by default or poly(ethylene oxide), poly(N-Vinylpyrrolidone), Tetrakis(4-carboxyphenyl)porphine, or other), polymer used in synthesis (none or poly(N-Vinylpyrrolidone), oleic acid, poly(ethylene oxide), BSA, or other), surfactant (none or L-ascorbic acid, ethylene glycol, sodium citrate, cetrimonium bromide, citric acid, trisodium citrate, ascorbic acid, or other), molar mass, Michaelis constant Km, molar maximum reaction rate Vmax, reaction type (substrate + co-substrate) (TMB + H2O2, H2O2 + TMB, TMB, ABTS + H2O2, H2O2, OPD + H2O2, H2O2 + GSH, or other), minimum concentration of the substrate when measuring catalytic activity C_min (mM), maximum concentration of the substrate when measuring catalytic activity C_max (mM), concentration of the co-substrate when measuring the catalytic activity (mM), concentration of nanoparticles in the measurement of catalytic activity (mg/mL), pH at which the catalytic activity was measured, and temperature at which the research was carried out (°C). You need to find all the experiments with different values mentioned in the article and write about each of them separately. It is imperative that each of these elements is addressed independently for every experiment, providing a complete and isolated description with accurate measurements in appropriate units. This approach will ensure a comprehensive and clear understanding of each experiment as an individual entity within the scientific literature on nanozymes. You should describe the experiments in the article separately in words while keeping the numerical values in the correct units of measurement. It is critically important to extract all the numerical values, as in the example, with particular emphasis on formula, activity, crystal system, length, width, depth (size or diameter), Km, Vmax, and reaction type. Usually, parameters such as Michaelis constant Km (mM) and Vmax (mM/s) are obtained in two experiments for every type of nanoparticle. You must determine which type of reaction these parameters belong to. The reaction type is H2O2 + TMB when H2O2 is a substrate and TMB is a co-substrate. The reaction type is TMB + H2O2 when TMB is a substrate and H2O2 is a co-substrate. For example, in the pair H2O2 and TMB: In the first case (Reaction type TMB + H2O2), H2O2 plays the role of a co-substrate with constant concentration (C_const, mM), and TMB plays the role of a substrate with concentrations ranging from C_min (mM) to C_max (mM). In the second case (Reaction type H2O2 + TMB), TMB plays the role of a co-substrate with constant concentration (C_const, mM), and H2O2 plays the role of a substrate with concentrations ranging from C_min (mM) to C_max (mM). Please divide all the data into two tracks: one where H2O2 was a substrate and its concentration varied, and one where H2O2 was a co-substrate and had a constant concentration. Please provide data only for those nanoparticles for which the kinetic assay was performed. All other parameters from the example are equally important. To answer questions, you can use the following tools: get_full_text, analyze_images, and llm_extractor. If you were unable to obtain the necessary information using the tools, mention that in your final answer. Do not attempt to use pre-existing knowledge. Do not use tools unnecessarily. After receiving a response from the tool, be sure to document your reasoning. Before writing the final answer, remember to write ‘Final answer:’.
Named Entity Recognition
The Named Entity Recognition agent processes the entire scientific article, divided into segments of 2048 tokens each. After each chunk undergoes processing, the agent generates a dictionary as a JSON object containing classes and their corresponding values for the given segment. In the subsequent merging step, we combine these dictionaries into a single comprehensive dictionary for the entire article. First, each dictionary is tagged with the chunk ID and timestamp of extraction, which helps track the source segment. Then, a simple conflict-resolution strategy is applied: if multiple dictionaries contain values for the same parameter class, the system checks whether these values are identical or differ only by formatting (e.g., slight textual variations). If multiple valid entries exist, all are retained under the same class with a note indicating their respective chunk IDs; if duplicates are detected, only one is kept. Finally, this merged dictionary is handed off to the main GPT-4o agent for holistic data interpretation and final output. Once all these dictionaries are combined into a comprehensive dictionary representing the entire article, it is fed into the main GPT-4o agent for a holistic information extraction analysis. The JSON dictionaries generated for each segment provide a structured representation of the relevant information, making it easier for the main GPT-4o agent to analyze and interpret the data. This agent serves as an additional expert, providing a unified perspective on the extracted information from the article as a whole.
NER agent training and hyperparameter tuning
We used pre-trained language models (PLMs) for further fine-tuning in our system. We carefully tuned the accumulation step, warmup steps, optimizer, and learning rate to achieve the best results with a Tree-structured Parzen Estimator (TPE)38. A Bayesian optimization technique aims to find the optimal set of hyperparameters that maximizes (or minimizes) an objective function. The accumulation step determines the number of gradients combined before updating the model weights, which can help stabilize training and reduce memory consumption39. Warmup steps gradually increase the learning rate during the initial training phase, preventing the model from diverging due to large gradients40.
Llama-3-8B was fine-tuned using a cross-entropy loss function, and the training process was carefully monitored to ensure convergence and optimal performance. We employed a learning rate scheduler with a linear warmup period followed by cosine annealing, which has been shown to improve generalization37. As shown in Fig. S1A, the warmup period was set to 334 steps, with an initial learning rate of 1.4e-6, gradually increasing to a maximum of 3e-6. The learning rate was then annealed according to the cosine annealing schedule over the remaining training steps. We used the AdamW41 optimizer with a weight decay of 0.01 and a batch size of 1. The accumulation step was set to 16, allowing for more efficient use of GPU memory while maintaining a larger adequate batch size. Figure S1B illustrates the learning rate schedule and the corresponding training loss throughout training. As shown in Figure S1C, the model achieved convergence after approximately 1,500 steps, with the validation loss stabilizing satisfactorily.
Mistral-7B was fine-tuned similarly but with different hyperparameters. Specifically, it was also trained using cross-entropy loss. Still, it employed approximately 368 warmup steps (Fig. S2). The initial learning rate was set to 5e-7, and the training process utilized a batch size of 4 with an accumulation step of 4. Additionally, we employed the Adafactor optimizer42 for the fine-tuning process. These adjustments in the training configuration allowed for a more tailored approach to fine-tuning the Mistral-7B model, potentially impacting its performance and adaptation to the specific task at hand.
Graph processing detection
GPT-4o allows for integrating and analyzing textual and visual information extracted from PDF documents, providing comprehensive results. To assist with image recognition, we employed the YOLOv8 model27. It is an advanced object detection algorithm that excels at detecting objects within an image in a single forward pass of the network.
The dataset used for training consisted of 537 annotated images from 50 scientific articles, covering various application classes: “table,” “figure,” and “scheme”. The images were manually labeled with object detection annotations for these three categories, ensuring a well-balanced dataset across them. The model was designed to detect each class effectively, even under challenging conditions such as overlapping elements, low resolution, or inconsistent formatting. The YOLOv8 model was trained for 250 epochs using a custom loss function that accounts for bounding box regression (box_loss), classification accuracy (cls_loss), and distribution-focused learning (dfl_loss). The training process was optimized using gradient descent with a scheduled learning rate and multi-scale data augmentation. The loss curves illustrate the model’s convergence during training, with the training and validation losses for box, classification, and distribution-focused objectives decreasing consistently across epochs. In particular, the training box loss and classification loss showed significant reductions, stabilizing around 0.2 and 0.17 by the end of training (epoch 250). The validation losses followed a downward trend, confirming that the model generalized well to unseen data (Figure S3). The trained YOLOv8 model was then tested on a variety of scientific documents. It accurately identified schemes, figures, and tables, providing bounding boxes and class labels as outputs (for metrics, see YOLO Training and Evaluation SI section). For instance, Figure S4 showcases an example where the model accurately detects and distinguishes between a scheme and a figure, even when presented nearby.
Integration of text and images
GPT-4o can automatically link textual descriptions with corresponding images and graphs, offering a complete data view. This is particularly important for analyzing complex scientific articles where visual data complements textual information.
Data extraction and interpretation
GPT-4o can extract critical parameters and trends from graphs and images, interpreting them in the context of the article. For example, it can recognize and analyze graphs representing the catalytic activity of nanozymes relative to their sizes, automatically extracting the data.
Structured data output
In the final stage of our system, we employ an automated process for converting extracted experimental data into structured JSON and tabular formats utilizing OpenAI’s structured outputs feature43. This process begins with a precisely crafted prompt instructing the language model to extract essential experimental parameters from scientific texts, including chemical formulas, catalytic activities, crystal systems, and catalytic constants. The prompt provides detailed guidelines on unit conversions, standardization of chemical formulas, and classification of crystal systems into predefined categories, ensuring consistency and precision across the extracted data. Once the data is extracted, it is organized into a machine-readable JSON format, with each parameter systematically mapped to its corresponding key. This structured output is then aggregated into a comprehensive table, facilitating subsequent analysis and database integration. By automating these stages, our system significantly reduces manual effort and ensures that new experimental data can be consistently and efficiently incorporated into the DiZyme knowledge base. Looking forward, nanoMINER holds the potential to fully automate the expansion of the DiZyme database, enabling it to continuously grow and adapt to the rapidly advancing field of nanozyme research without the limitations imposed by traditional manual curation processes. After generating the table, we validated the results by comparing them with the reference dataset using the Jaccard index44 to assess the precision of the extraction.
System evaluation
Our evaluation methodology focuses on complete experiment extraction at the paper level, using custom precision and recall calculations specifically designed for our extraction task. To evaluate our extraction system’s performance, we compared automatically extracted parameters against a manually annotated test dataset containing papers with 1-6 experiments each. The evaluation was conducted at multiple levels. For each paper, we ran the extraction process 100 times to assess consistency, as language models may produce slight variations in outputs. We set the temperature to 0 for all large language model components to encourage deterministic behavior. However, we note that when using OpenAI APIs, even with a fixed temperature of 0, minor variations in outputs can occur across runs. These small inconsistencies likely stem from non-deterministic system-level factors such as tokenization differences, API backend sharding, and minor sampling randomness. As a result, we performed 100 repeated extraction runs to estimate the mean and variability of performance metrics. We calculated precision and recall for each parameter type across all experiments within a paper.
Precision
For each paper in our test dataset, the Precision is calculated as the ratio of correctly extracted experiments to the total number of experiments extracted by the system. An experiment is correctly extracted when all its parameters match the ground truth values. Then, the average precision was calculated using all papers in the test dataset:
Recall
For each paper, the Recall is calculated as the ratio of correctly extracted experiments to the total number of experiments present in the ground truth for that paper. Then, the average recall was calculated using all papers in the test dataset:
Levenshtein distance
In addition to precision and recall, we employed the Levenshtein distance as a metric for evaluating the similarity between string-based experimental parameters. The Levenshtein distance measures the minimum number of insertions, deletions, or substitutions required to transform one string into another. It is well-suited for comparing textual data such as chemical formulas, polymer types, or crystal systems. For our analysis, we utilized the normalized Levenshtein distance, which is calculated as:
Here \(s1\) and \(s2\) are the two strings being compared. This metric produces a value between 0 and 1, where 0 indicates perfect similarity, and 1 indicates maximum dissimilarity. This allows us to rigorously assess the accuracy of our language model’s output by comparing extracted parameters with the reference ground truth from scientific texts.
For example, consider two similar strings, “CuZnFeS” and “Cu5ZnFe5S6”. The normalized Levenshtein distance between these two strings is:
This score reflects a small degree of dissimilarity between the two strings. By incorporating the normalized Levenshtein distance, we systematically evaluate the accuracy of string-based data extractions across our dataset, ensuring robust and precise text processing in our automated system.
Jaccard Index
The Jaccard index is a widely utilized metric for quantifying the similarity between two sets, particularly in tasks involving data comparison and extraction41,42. It is calculated as the ratio of the size of the intersection of the sets to the size of their union and is mathematically expressed as an equation:
where \({|A}\cap {B|}\) denotes the number of common elements between sets A and B and \({|A}\cup {B|}\) represents the total number of unique aspects across both sets. The Jaccard index measures the similarity between two sets, with a value between 0 and 1. It is sensitive to even minor differences and tends to be conservative, with values close to 1 indicating near-perfect alignment.
The Jaccard index serves as a robust metric for evaluating the accuracy of our large language model-based system in extracting experimental data from nanozyme-related literature. To elucidate the practical application of this index, consider a scenario where a scientific paper delineates ten experiments, and our automated system accurately identifies 8 out of 10 values for a specific parameter. The Jaccard index would be computed as follows:
This score of 0.67 reflects the system’s high success rate while simultaneously demonstrating the index’s stringency in penalizing even minor discrepancies. The Jaccard index’s formulation ensures it accounts for correctly identified and missing values, providing a comprehensive performance measure. To further illustrate the index’s sensitivity, consider an alternative scenario where 2 out of 3 values are correctly extracted. In this case, the Jaccard index would be:
Data availability
The data and the code supporting this study’s findings are available at https://github.com/ai-chem/nanoMINER.git.
References
Foppiano, L., Lambard, G., Amagasa, T. & Ishii, M. Mining experimental data from materials science literature with large language models: an evaluation study. Sci. Technol. Adv. Mater. Methods 4, 2356506 (2024).
Fine-tune BERT with Sparse Self-Attention Mechanism - ACL Anthology. https://aclanthology.org/D19-1361/ (accessed 2024-06-20).
Guo, M.-H. et al. Attention mechanisms in computer vision: a survey. Comput. Vis. Media 8, 331–368 (2022).
Zhang, H., Goodfellow, I., Metaxas, D., Odena, A. Self-Attention Generative Adversarial Networks. In Proceedings of the 36th International Conference on Machine Learning; PMLR, 2019; 7354–7363.
Li, W., Qi, F., Tang, M. & Yu, Z. Bidirectional LSTM with self-attention mechanism and multi-channel features for sentiment classification. Neurocomputing 387, 63–77 (2020).
Peng, R., Liu, K., Yang, P., Yuan, Z., Li, S. Embedding-based retrieval with LLM for Effective Agriculture Information Extracting from Unstructured Data. arXiv 2023. https://doi.org/10.48550/ARXIV.2308.03107.
Patiny, L., Godin, G. Automatic Extraction of FAIR Data from Publications Using LLM. ChemRxiv December 4. https://doi.org/10.26434/chemrxiv-2023-05v1b-v2. (2023)
Biswas, A. & Talukdar, W. Robustness of structured data extraction from in-plane rotated documents using multi-modal Large Language Models (LLM). J. Artif. Intell. Res. 4, 176–195 (2024).
Jiang, A. Q. et al. Mistral 7B. arXiv. https://doi.org/10.48550/ARXIV.2310.06825. (2023)
Introducing Meta Llama 3: The most capable openly available LLM to date. https://ai.meta.com/blog/meta-llama-3/ (accessed 2024-06-20).
Radford, A., Wu, J., Child, R., Luan, D., Amodei, D. & Sutskever, I. Language models are unsupervised multitask learners. OpenAI Tech. Rep. (2019).
Evaluating GPT-4V (GPT-4 with Vision) on Detection of Radiologic Findings on Chest Radiographs | Radiology. https://doi.org/10.1148/radiol.233270 (accessed 2024-06-20).
Sun, Q., Cui, X., Zhou, W., Li, H. Exploiting GPT-4 Vision for Zero-Shot Point Cloud Understanding. arXiv January 15, 2024. https://doi.org/10.48550/arXiv.2401.07572.
OpenAI. GPT-4o System Card. August 8, 2024. https://cdn.openai.com/gpt-4o-system-card.pdf.
Wu, Y., Hu, X., Fu, Z., Zhou, S., Li, J. GPT-4o: Visual perception performance of multimodal large language models in piglet activity understanding. arXiv June 14, 2024. https://doi.org/10.48550/arXiv.2406.09781.
Schilling-Wilhelmi, M. et al. From text to insight: large language models for materials science data extraction. arXiv July 23, 2024. http://arxiv.org/abs/2407.16867 (accessed 2024-09-09).
Wang, X., Huey, S. L., Sheng, R., Mehta, S., Wang, F. SciDaSynth: Interactive structured knowledge extraction and synthesis from scientific literature with large language model. arXiv April 21, 2024. http://arxiv.org/abs/2404.13765 (accessed 2024-09-09).
Dagli, M. M. et al. Development and validation of a novel AI framework using NLP with LLM integration for relevant clinical data extraction through automated chart review. Sci. Rep. 14, 26783 (2024).
Autonomous Discovery in the Chemical Sciences Part II: Outlook - Coley - 2020 - Angewandte Chemie International Edition - Wiley Online Library. https://doi.org/10.1002/anie.201909989 (accessed 2024-06-20).
Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Central Science. https://doi.org/10.1021/acscentsci.9b00576 (accessed 2024-06-20).
The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminform. | Full Text. https://doi.org/10.1186/1758-2946-7-S1-S2 (accessed 2024-06-20).
Dagdelen, J. et al. Structured information extraction from scientific text with large language models. Nat. Commun. 15, 1418 (2024).
Sun, L. et al. ChatGPT combining machine learning for the prediction of nanozyme catalytic types and activities. J. Chem. Inf. Model. 2024, acs.jcim.4c00600. https://doi.org/10.1021/acs.jcim.4c00600.
Lee, S. et al. Data-driven analysis of text-mined seed-mediated syntheses of gold nanoparticles. Digit. Discov. 4, 93–104 (2025).
Ansari, M. & Moosavi, S. M. Agent-based learning of materials datasets from the scientific literature. Digit. Discov. 3, 2607–2617 (2024).
Solovev, G. V. et al. Towards LLM-driven multi-agent pipeline for drug discovery: neurodegenerative diseases case study. In Int. Conf. Learn. Represent. (ICLR) (2024).
Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); IEEE: Las Vegas, NV, USA, 2016; 779–788. https://doi.org/10.1109/CVPR.2016.91.
Yao, S. et al. ReAct: Synergizing Reasoning and Acting in Language Models. arXiv 2022. https://doi.org/10.48550/ARXIV.2210.03629.
Comprehensive Examination of Instruction-Based Language Models: A Comparative Analysis of Mistral-7B and Llama-2-7B|IEEE Conference Publication | IEEE Xplore. https://ieeexplore.ieee.org/abstract/document/10434081 (accessed 2024-06-20).
Razlivina, J., Dmitrenko, A. & Vinogradov, V. AI-powered knowledge base enables transparent prediction of nanozyme multiple catalytic activity. J. Phys. Chem. Lett. 15, 5804–5813 (2024).
Chataut, S. et al. Comparative study of domain driven terms extraction using large language models. arXiv 2024. https://doi.org/10.48550/ARXIV.2404.02330.
Mimicking horseradish peroxidase and oxidase using ruthenium nanomaterials - RSC Advances (RSC Publishing). https://pubs.rsc.org/en/content/articlelanding/2017/ra/c7ra10370k.
Hiroki Nakayama and; Takahiro Kubo and; Junya Kamura and; Yasufumi Taniguchi and; Xu Liang. doccano: Text Annotation Tool for Human. doccano. https://doccano.web.webis.de/ (accessed 2024-06-20).
Decker, J. pdfplumber: Plumb a PDF for detailed information about each char, rectangle, line, etc. GitHub. https://github.com/jsvine/pdfplumber (2023).
Papers with Code - PaDeLLM-NER: Parallel Decoding in Large Language Models for Named Entity Recognition. https://paperswithcode.com/paper/padellm-ner-parallel-decoding-in-large (accessed 2024-06-20).
Paolini, G. et al. Structured prediction as translation between augmented natural languages. In Int. Conf. Learn. Represent. (ICLR) 1–20 (2021).
Devlin, J., Chang, M.-W., Lee, K., Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv May 24, 2019. https://doi.org/10.48550/arXiv.1810.04805.
Watanabe, S. Tree-structured parzen estimator: understanding its algorithm components and their roles for better empirical performance. arXiv May 26, 2023. http://arxiv.org/abs/2304.11127 (accessed 2024-06-20).
Ott, M. et al. Fairseq: A fast, extensible toolkit for sequence modeling. arXiv April 1, 2019. https://doi.org/10.48550/arXiv.1904.01038.
Popel, M. & Bojar, O. Training Tips for the Transformer Model. Prague Bull. Math. Linguist. 110, 43–70 (2018).
Loshchilov, I., Hutter, F. Decoupled weight decay regularization. arXiv January 4, 2019. https://doi.org/10.48550/arXiv.1711.05101.
Shazeer, N., Stern, M. Adafactor: Adaptive learning rates with sublinear memory cost. arXiv April 11, 2018. https://doi.org/10.48550/arXiv.1804.04235.
Pokrass, M. Introducing Structured Outputs in the API. https://openai.com/index/introducing-structured-outputs-in-the-api/ (accessed 2024-08-10).
Hancock, J. M. Jaccard Distance (Jaccard Index, Jaccard Similarity Coefficient). In Dictionary of Bioinformatics and Computational Biology; Hancock, J. M., Zvelebil, M. J., Eds.; Wiley, 2004. https://doi.org/10.1002/9780471650126.dob0956.
Acknowledgements
The work was financially supported by the Priority 2030 Federal Academic Leadership Program. We would like to thank S. Danilova, M. Reykina, and S. Shevtsova for their valuable contribution to the annotation of the dataset used in this study.
Author information
Authors and Affiliations
Contributions
R.O. designed and implemented a multi-agent system with advanced prompting. K.R. was responsible for training and testing the LLM agent and compiling the article. S.M. participated in selecting articles for training and annotating them, writing prompts, and evaluating model responses. O.Z. was responsible for experiment planning, task allocation within the team, and developing baseline solutions for LLM tasks. R.S. integrated with the DiZyme service and integrated LLM with an interface on Streamlit. R.K. was responsible for ensuring the accurate parsing of PDF documents and analyzing images within the articles. J.R. supervised the research, annotating data and evaluating the model. J.R., K.R., S.M., O.Z., R.K., R.O., R.S., A.D. wrote the paper. A.D. and V.V. conceived the project and supervised the work. All authors contributed to and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Odobesku, R., Romanova, K., Mirzaeva, S. et al. Agent-based multimodal information extraction for nanomaterials. npj Comput Mater 11, 194 (2025). https://doi.org/10.1038/s41524-025-01674-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41524-025-01674-7
This article is cited by
-
The emerging role of machine learning in nanomaterials research: applications, challenges, and future directions
Journal of Nanoparticle Research (2025)