Abstract
Microgravity affects cell growth and apoptosis. Using a random positioning machine, we previously found that simulated microgravity (SMG) suppresses total cell proliferation and induces apoptosis of in vitro cultured hepatocytes. Here, we report an interesting finding that SMG partially suspends these hepatocytes, forming large spheroids. Both attached and suspended cells can grow, but suspended cells exhibit reduced cell growth and apoptosis, implying dormancy. Mechanistically, attached but not suspended cells exhibit upregulated PI3K/AKT/mTOR pathway activity and downstream protein c-Myc. Knocking down c-Myc promotes, whereas overexpressing it suppresses, hepatocyte suspension under SMG, suggesting that the PI3K/AKT/mTOR pathway and c-Myc protect these cells from entering a dormant state upon SMG.
Similar content being viewed by others
Introduction
Living organisms on the surface of the earth are influenced by gravity. However, during spaceflight, astronauts experience microgravity, which significantly impacts cellular and tissue functions1. The liver is a vital organ with various physiological functions2, which can be disrupted by microgravity. For example, long-term spaceflight/simulated microgravity (SMG) affects hepatocyte growth and proliferation3,4,5,6. In agreement with these findings, rats exposed to nine days of spaceflight exhibited reduced hepatic production of heat shock protein 90 and the cell cycle regulator p537; mice subjected to 12 days of spaceflight showed increased expression of genes related to hepatocyte differentiation and apoptosis8. Also, rats placed in a tail-suspension position for 6 weeks, which simulates microgravity, exhibited inhibited hepatocyte proliferation, which was indicated by decreases in Ki67 and several cyclins and cyclin-dependent kinases9. Transcriptomic and metabolomic analyses related accumulation of oxidized proteins and mitochondrial dysfunction to microgravity, which may trigger hepatocyte senescence and steatosis10.
Phosphatidylinositide 3-kinases (PI3Ks) and their downstream proteins Ser and Thr kinase (AKT) and mammalian target of rapamycin (mTOR) play crucial roles in cell proliferation, survival, and metabolism11. PI3K/AKT/mTOR activation upregulates MYC12, transcriptional factors that promote cell growth13,14,15, and its dysregulation is implicated in multiple cancers, such as hepatocellular carcinoma16. Indeed, c-Myc plays a key role in hepatocyte transition from the G0/G1 to S phase17. However, whether the PI3K/AKT/mTOR pathway and c-Myc play a role in microgravity-mediated hepatocyte growth defects is unclear. Interestingly, long-term spaceflight activates the PI3K signaling pathway which may related to c-Myc in human fibroblasts18 and upregulates c-Myc in mouse ventricles19, suggesting that similar events may occur in hepatocytes.
Previously, we found that SMG suppresses hepatocyte growth, induces apoptosis, and disrupts cellular functions20. Here, we further study that SMG causes some hepatocytes to form suspending clusters, which shown different morphological and genomic characteristics to attached cells. Combined, our data identified a novel cellular response to SMG stress, in which c-Myc plays a key role.
Results
SMG disrupts hepatocyte morphology, growth, and viability
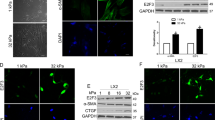
Using a random positioning machine (RPM) to simulate microgravity, we previously showed that SMG suspends hepatocytes in a time-dependent manner, disrupts NCTC1469 hepatocyte proliferation and induces apoptosis20. However, the precise mechanism is unclear. Under NG conditions for three days (Fig. 1A), cultured NCTC1469 hepatocytes attach to the well bottom and form a monolayer in vitro (Fig. 1B); however, SMG causes 75% of cells to form multicellular spheroids suspended in the media in three days (Fig. 1C), which could explain SMG-induced proliferation and apoptotic defects. Here, we call these cells SMG-suspending (SMG-S) cells and the remaining attached cells SMG-attached (SMG-A) cells. Then, we counted the number of cells treated with SMG for three days and found that this treatment significantly decreased cell numbers of SMG-S and SMG-A, compared to NG conditions (Fig. 1D). Consistent with these data, SMG for three days significantly decreased the nuclear protein Ki67 both in SMG-S and SMG-A cells (Fig. 1E), suggested lower proliferative potentials.
A Experimental procedure. B Microscopic appearance of NCTC cells after three days of culture under NG and SMG conditions (scale bar: 50 μm). C Cell number of NCTC cells after three days of culture under NG and SMG conditions. D, E Ki67 staining analysis of NCTC cells apoptosis after three days of culture under NG and SMG conditions (scale bar: 100 μm). Results are presented as mean ± SEM, Statistical analysis was performed using one-way ANOVA followed by Tukey’s tests, *p < .05, **p < .01, ***p < .005, N = 3.
SMG-A and SMG-S cells exhibit different growth and death defects
To further assess the proliferation and apoptosis of SMG-S and SMG-A cells, we performed flow cytometry analyses. As shown in Fig. 2A, B, for SMG-S cells, the cell proportions of both G0/G1, S and G2/M phase were unaltered, suggesting no change in cell cycle progression. Conversely, for SMG-A cells, the proportions of G0/G1 and S phase cells were decreased and increased respectively, suggesting a cell cycle arrest. Importantly, the proportion of sub-G1 phase cells is increased for SMG-A cells, suggesting more apoptosis, which is further confirmed by Annexin V-FITC and propidium iodide (PI) staining (Fig. 2C–F). Annexin V-FITC and PI stain early and late stages of apoptotic cells, respectively. Interestingly, the SMG-S group exhibited more FITC-positive and PI-negative cells, whereas the SMG-A group exhibited more FITC- and PI-positive cells, compared to NG, suggesting that SMG-S and SMG-A groups respectively have more early and late apoptosis. Thus, apoptosis may be delayed or impaired in SMG-S cells.
A, B Distribution of each phase of the cell cycle was evaluated in NCTC cells after three days of culture under NG and SMG conditions. C–F Annexin V-FITC staining analysis of NCTC cells proliferation after three days of culture under NG and SMG conditions. Results are presented as mean ± SEM, Statistical analysis was performed using one-way ANOVA followed by Tukey’s tests, *p < 0.05, **p < 0.01, ***p < 0.005, N = 3.
In view of the difference of apoptotic ratio is very small. Next, we assessed the proliferating propensity of SMG-S and SMG-A cells. After collecting these cells, we replated and cultured them under NG for three days and counted cell numbers. As shown in Supplementary Fig. 1, SMG-S but not SMG-A cells grow more slowly than the control, suggesting that SMG-S exhibits reduced cytoactive. In summary, our data suggested that SMG-A cells exhibit cell cycle arrest and increased apoptosis, whereas SMG-S cells exhibit reduced cytoactive.
Cell-cycle-related genes are differentially expressed in SMG-S and SMG-A cells
Next, we investigated the mechanism by which SMG causes defects in cell growth and apoptosis. In a previous study, we compared the transcriptomes of SMG (including both SMG-A and SMG-S) and NG hepatocytes and identified that most differentially expressed genes (DEGs) are associated with lipid metabolism, inflammatory response, immune response, and cell cycle arrest20. Here, we performed RNA sequencing (RNA-seq) analyses and compared the transcriptomes of SMG-S and SMG-A cells, followed by western blot and real-time (RT)-PCR (Fig. 3A). As shown in Fig. 3B, we identified 1178 DEGs, with 496 downregulated and 682 upregulated in SMG-S cells, compared to SMG-A (Fig. 3B). Interestingly, gene ontology (GO) enrichment analyses suggested that most downregulated DEGs play roles in DNA replication and repair, cell cycle, etc. (Fig. 3C), consistent with that SMG-S cells exhibit reduced growth activity. In addition, most up-regulated DEGs are implicated in immune responses (Fig. 3C), implying a role of immunity in cells responding to SMG.
A Experimental procedure for RNA sequencing. B Volcano plot of DEGs in NCTC cells after three days of culture under NG and SMG conditions. C GO-BP enrichment analysis of NCTC cells after three days of culture under NG and SMG conditions. D qPCR validation of gene expression in NCTC cells after three days of culture under NG and SMG conditions. E Western blot validation of protein expression in NCTC cells after three days of culture under NG and SMG conditions. Results are presented as mean ± SEM, Statistical analysis was performed using one-way ANOVA followed by Tukey’s tests, *p < 0.05, **p < 0.01, ***p < 0.005, N = 3.
Next, we compared the transcriptomes of the SMG-A and SMG-S cells versus NG. As shown in Supplementary Fig. 2, most upregulated DEGs in both SMG-A and SMG-S cells are implicated in immune responses, and most downregulated DEGs in both groups were implicated in DNA replication and repair and cell cycle. These findings further support that SMG disrupts cell growth and immunity.
Interestingly, we noted that c-Myc and several of its downstream genes, e.g., Cyclin D (Ccnd), Cyclin E (Ccne), Mcm were among the downregulated DEGs. C-Myc is required for the G0/G1-S transition in primary hepatocytes17, which plays role in regulating CCND2 expression21. To verify our RNA-seq data, we performed RT-PCR and western blot assay. As shown in Fig. 3D and E, the mRNA and/or protein levels of c-Myc and Ccnd2 in SMG-S cells were much less than in SMG-A cells. Interestingly, the mRNA/protein levels of these proteins in SMG-S were unchanged compared to NG, but in SMG-A, they were strongly upregulated. Meanwhile, the mRNA levels of Ccne1, Mcm2, Mcm4, and Mcm5 were significantly downregulated in SMG-S compared to SMG-A and NG (Supplementary Fig. 3). Combined, our data suggested that SMG-A, but not SMG-S cells exhibit upregulated c-Myc activity.
PI3K/AKT/mTOR pathway was downregulated in SMG-S compared to SMG-A
Next, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses on SMG-S versus SMG-A DEGs. As shown in Fig. 4A, we found that downregulated DEGs enriched in genes related to cell cycle progression, the p53, PI3K-AKT, etc. signaling pathway, and DNA replication. We also performed gene set enrichment analysis (GSEA) of KEGG pathways. This analysis indicated that down-regulated DEGs were enriched in genes related to cell cycle and DNA replication, etc. (Fig. 4B). To confirm these findings, we first performed immunostaining on NCTC cells and showed that the p53 level is reduced in SMG-S cells, compared to SMG-A (Fig. 4C, D). Additionally, we further performed western blot assay of p53 and p53-dependent apoptosis protein expression. We found that the protein levels of p53, BCL2 and FAS are reduced in SMG-S cells, compared to SMG-A, as for BAX, was not significant affected in these two groups (Fig. 4E, F). Next, we assessed the activity of the PI3K/AKT/mTOR pathway. As shown in Fig. 4G and H, phosphorylated PI3K, AKT, and mTOR are less in SMG-S cells compared to SMG-A, suggesting reduced pathway activity in SMG-S cells. Similar to c-Myc and Ccnd2, phosphor-PI3K, AKT, and mTOR are mostly upregulated in SMG-A, but not SMG-S, cells, implying a correlation of c-Myc and PI3K/AKT/mTOR pathway activity22.
A KEGG pathway analysis of NCTC cells after three days of culture under NG and SMG conditions. B GSEA of NCTC cells after three days of culture under NG and SMG conditions. C, D p53 staining analysis of NCTC cells apoptosis after three days of culture under NG and SMG conditions (scale bar: 20 μm). E–H Western blot validation of protein expression in NCTC cells after three days of culture under NG and SMG conditions. Results are presented as mean ± SEM, Statistical analysis was performed using one-way ANOVA followed by Tukey’s tests, *p < 0.05, **p < 0.01, ***p < 0.005, N = 3.
c-Myc plays a key role in SMG-A/SMG-S transition
To investigate whether c-Myc plays a role in the transition from the SMG-A to SMG-S state, we performed c-Myc knockdown and overexpression respectively using its siRNA and cDNA (Fig. 5A). We used three independent siRNA lines and found that two of them (#1 and 3) significantly reduced c-Myc as well as downstream p53 expression and increased the ratio of SMG-S versus SMG group total cells (Fig. 5B–D). In addition, c-Myc overexpression reduces this ratio (Fig. 5E–H). Hence, a high c-Myc level seems to maintain cells attached under SMG.
A Experimental procedure for c-Myc knockdown and overexpression. B Verification of c-Myc knock-down by siRNA. C Microscopic appearance of NCTC cells treated with siRNA after three days of culture under NG and SMG conditions (scale bar: 50 μm). D Quantification of suspending cell number after siRNA treatment. E Transfection efficiency of c-Myc over-expressing plasmid indicated by fluorescence (scale bar: 100 μm). F Verification of c-Myc over-expressing level. G Microscopic appearance of the NCTC cells treated with plasmid after three days of culturing under NG and SMG conditions (scale bar: 50 μm). H Quantification of suspending cell number after over-expressing treatment. Results are presented as mean ± SEM, Statistical analysis was performed using T test and one-way ANOVA followed by Dunnett’s test, *p < 0.05, ***p < 0.005, N = 3.
To further verify the role of PI3K/AKT/mTOR pathway and c-MYC in SMG-A/SMG-S transition, we applied 10 μM AKT inhibitor (LY294002) and c-MYC inhibitor (10058-F4) respectively. As shown, the spheroids are significant more when two inhibitors used under SMG for 3 days (Supplementary Fig. 4).
Discussion
Previous studies have shown that SMG exposure significantly decreased proliferation and increased apoptosis in various cell types3,4,5,6. Our findings suggest SMG disrupts hepatocyte growth and survival. Interestingly, the suspending (SMG-S) cells are alive but exhibit reduced cell proliferation and apoptosis, compared to attached (SMG-A) cells, suggesting reduced cell activity. Mechanistically, the PI3k/AKT/mTOR pathway is hyperactive, and its downstream protein c-Myc is upregulated, in SMG-A but not SMG-S cells. Furthermore, overexpressing c-Myc inhibits, and knocking down it promotes hepatocyte suspension. Our transcriptome analysis revealed that some key DEGs associated with cell cycle regulation, such as c-Myc and CCND2, all of which are crucial for G1/S transition and overall cell cycle progression17,23,24,. The disruption of these genes under SMG conditions likely leads to cell cycle arrest and reduced proliferation. Additionally, the increase in apoptosis may be linked to stress responses activated by the altered extracellular matrix, which could trigger cell death pathways25.
One of the interesting findings of our study was the transition of hepatocytes from SMG-A to SMG-S under SMG conditions. The transition of prostate cancer cells to a suspension state and the associated differential expression of the cytokines IL-1α, IL-1β, IL-6 and IL-8, also supporting SMG involvement in cellular state transition26. It suggests a profound alteration in cell adhesion properties, which are vital for maintaining tissue architecture and function25. The loss of adhesion is often associated with changes in the expression of cell adhesion molecules and the extracellular matrix, as well as alterations in cytoskeletal dynamics25.
Our previous whole transcriptome analysis revealed significant changes in genes related to cell adhesion and the cytoskeleton20, supporting the observed phenotypic changes. Furthermore, the involvement of the PI3K/AKT/mTOR pathway was confirmed through RNA sequencing, RT-PCR, and western blot. This pathway is known to play a critical role in cell growth, survival, and metabolism11, thus its disruption under SMG conditions may contributes to the observed changes in cellular state transition. Meanwhile, it has been known that choroidal vascular endothelial cells exposed to SMG exhibit ultrastructure and apoptosis, which is related to phosphorylated forms of PI3K and Akt, enhanced along with the time of exposure to SMG27. In our study, SMG-A has a higher phosphorylation level of PI3K and Akt than NG and SMG-S, concomitantly exhibiting more apoptotic propensity.
Our data also showed that c-Myc expression is significantly altered under SMG conditions, hence manipulating c-Myc levels may have a profound effect on cell behavior. Specifically, c-Myc knockdown increased the number of cells in the suspension state, while c-Myc overexpression decreased it, suggesting that c-Myc plays a key role in maintaining the attached state of hepatocytes under SMG. MYC is a well-known transcription factor that regulate of cell proliferation15, growth17, and apoptosis28. The upregulation of c-Myc under SMG conditions likely contributes to the increased apoptosis observed in our study. Furthermore, the ability of c-Myc to modulate the transition between attached and suspension states underscores its crucial role in mediating cell adhesion and cytoskeletal dynamics29.
The relationship between p53 and c-Myc is well investigated. It has been reported that p53 is a target gene of c-Myc30,31. In our study, the protein expression of p53 in the cells was downregulated, correlated with c-Myc silencing. Additionally, we detected the p53-dependent apoptosis protein FAS, BAX and BCL-2. The expression of FAS and BCL2 is correlation with p53, otherwise the result of BAX is opposite. Thus we assume that the apoptosis is due to extrinsic apoptotic signals. However, the difference of apoptotic ratio is very small in the SMG group. Then we detected both the SMG-S and SMG-A cells potential possess proliferating propensity, which support from the side the ratio of apoptotic cells in the SMG group to some degree.
The findings from our study have significant implications for understanding the impact of microgravity on liver function during spaceflight. The liver plays a critical role in metabolism, detoxification, and protein synthesis, which may have severe consequences for astronaut health if in disorder32. The PI3K/AKT/mTOR pathway may interact with c-Myc to modulate cell cycle in many type of cells22,33. Furthermore, the paper has demonstrated that PI3K/Akt/mTOR pathway regulate the mtp53 expression via c-myc31. Our data suggest that targeting the PI3K/AKT/mTOR pathway and c-Myc could be a potential strategy for mitigating the adverse effects of microgravity on hepatocytes. The use of exogenous c-Myc inhibitors to reduce p53-dependent apoptosis could help maintain liver function during long-duration space missions. Additionally, pharmacological agents that modulate the PI3K/AKT/mTOR pathway could be explored as countermeasures to protect hepatocytes from the detrimental effects of microgravity.
While our study provides valuable insights into the molecular mechanisms underlying the response of hepatocytes to SMG, further research is needed to fully elucidate these pathways and their interactions. Investigating the upstream regulators and downstream effectors of the PI3K/AKT/mTOR pathway and c-Myc will enhance our understanding of how microgravity impacts cell behavior.
Methods
Cell Cultures and SMG exposure
The NCTC-1469 cell line was obtained were seeded into T12.5 cm2 flasks. The cells were cultured in RPMI 1640 medium (Gibco, Fisher Scientific, Schwerte, Germany) supplemented with 10% FBS (Sigma Aldrich, Steinheim, Germany) and 1% penicillin/streptomycin (Life Technologies, New York, United States). The culture medium was replaced every three days, and upon reaching 70–80% confluence, the cells were split at a 1:5 ratio. Care was taken to pour the medium into the flasks without creating bubbles to avoid fluid shearing.
The random positioning machine (RPM) used in this study consists of two independently rotating frames34. By continuously and randomly changing their direction, the gravity vector of Earth becomes negligible over time. The flasks were securely fixed in the RPM, while the control was maintained under 1 G conditions in the same CO2 incubator.
Cell number counting
Culture flasks containing 2 × 105 NCTC1469 cells were mounted on RPM for three days. The normal gravity (NG) were placed next to the RPM and cultured in parallel. Cell numbers were quantified using the automatic cell counter. Briefly, NCTC1469 cells (2 × 105/mL) were seeded into the T12.5 flask for 24 h to allow adherence. They were then mounted on a random positioning machine (RPM) for three days. The medium was collected and centrifuged at 200 × g to isolate the SMG-S cells. The attached cells from the NG and SMG-A were harvested using 0.25% Trypsin–EDTA. The cells were resuspended in 1 mL PBS, and 10 μL of this suspension was placed onto a Nanoentek C-C hip for automatic cell counting.
Immunofluorescent staining
At culturing for three days, the NCTC1469 cells in flasks under different culture conditions are collected and prepared for immunostaining. Cells were fixed with 4% paraformaldehyde solution at room temperature for 30 min, then permeabilized for 20 min with 0.2% Triton X-100 and donkey serum at room temperature and add the 5% donkey serum blocking solution for 1 h blocking at 37 °C. Incubated the cell in primary antibody solution for overnight at 4 °C, subsequently with the second antibody (1:200 dilutions) and Hoechst 33342 (0.1–1 μg/mL) in 5% donkey serum for 1 h at room temperature. Washed the cells twice for 5 min in PBS. Put one drop of mounting medium, add coverslip and seal with nail polish. Observed the slides and acquire the image under the confocal microscope.
Apoptotic analysis with annexin V
The annexin V/PI double staining kit was used to determine the different states of NCTC cells. The cells were washed twice with PBS and harvested by centrifugation. Pre- diluted annexin V (5 μL) with PBS (195 μL) and PI (10 μL) was added to the cells in dark. Half an hour later, cells in different apoptosis stages were analyzed within an hour by flow cytometry.
Cell cycle analysis
For cell cycle analyses by flow cytometry, cells were trypsinized, washed with PBS, and then fixed in 70% ice- cold ethanol overnight. Samples were rinsed twice with PBS after removal of ethanol. Propidium iodide (PI) staining solution (1% Triton X-1 00, 0.01% RNase, 0.05% PI) was added to the cell suspensions, and the mixtures were incubated for 30 min at 4 °C in dark to stain DNA. A flow cytometer was used for cell- cycle analysis, and for each sample, 10,000 events were recorded. Data acquisition and analyses were done using the CytExpert v2.4, and the percentage of cells present in the sub-G 1, G0/G1, S, and G2/M (mitosis) phases were determined.
Total RNA isolation and sequencing
Total RNA was isolated from the NG and SMG hepatocytes using Trizol Reagent (Life Technologies, Carlsbad, USA), according to the manufacturer’s protocol. Total RNA was quantified using the ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). The RNA integrity and absence of genomic DNA contamination were evaluated by agarose gel electrophoresis. The RNA samples were sequenced using a standard Illumina protocol by Origingene Co. Ltd. (Shanghai, China). Reads were mapped to the current Indian rhesus macaque reference genome (rheMac8) by HISAT2 software. Gene counts were estimated by HTSeq (0.11). The R package DESeq2 was applied to determine differentially expressed genes. Genes with a fold-change >2 and an adjusted p < 0.05 were defined as differentially expressed genes (DEGs). The R package top GO was applied to perform GO enrichment analysis of the DEGs. GSEA was performed according to the GSEA manual.
Quantitative real-time PCR (qPCR) analysis
NCTC 1469 cells were lysed with the RNeasy Plus Mini Kit, and the total mRNA was extracted according to the manufacturer’s protocol. cDNA synthesis was performed using the ReverTra Ace qPCR RT Master Mix with gDNA Remover. mRNA levels were measured using a qPCR System with the THUNDERBIRD SYBR qPCR Mix, according to the provided protocol. Target gene primers used in this study are listed in Supplementary Table 1. Relative gene expressions were analyzed using the comparative threshold cycle method (2−∆∆t) and normalized to hypoxanthine phosphoribosyltransferase (Hprt).
Western blot
The cells for western blot were cultured for three days in T12.5 (JET BIOFIL, Guangzhou, China) with about 36 ml of culture medium. Cells were trypsinized, collected and lysed for 30 min on ice in 100 μl of cold RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China), 1 ml of RIPA contains 10 μl of phenylmethanesulfonyl fluoride (PMSF, Beyotime Biotechnology, Shanghai, China), 5 μl of protease inhibitor cocktail solution and phosphatase inhibitor Phos-stop respectively (Sigma-Aldrich, St. Louis, USA). The lysate was followingly added with 25 μl 5× loading buffer (Beyotime Biotechnology, Shanghai, China), then 10 μl of samples were resolved by PAGE (Criterion 4–20% Tris-HCl; Applygen, Beijing, China). Proteins were blotted onto PVDF membrane (Merck Millipore, Darmstadt, Germany), and the blot was then incubated in blocking buffer (5% nonfat BSA in 10 mM Tris-HCl, 1.15 M NaCl, and 0.1% Tween-20) for 1 h at room temperature. Immunoblotting entailed the following: incubation overnight with those primary antibodies at 4 °C (listed in Supplementary Table 2) followed with 2 h incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies. The labeling was visualized using the chemiluminescence HRP substrate (Thermo Fisher Scientific, USA). In each experiment, band densities were normalized against HPRT, and the results are expressed as protein level to control.
RNAi of c-Myc
Three pairs of interference sequences that targeted mouse c-Myc mRNA (siRNA1, siRNA2, siRNA3) and the non-specific control siRNA (siCtrl) were synthesized by Ruibobio (Guangzhou, China) and their sequences are listed in Supplementary Table 3. Following cell seeding for 24 h, NCTC1469 cells were transfected with 12.5 nM siRNAs respectively using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 48 h transfection, cells were transferred to RPM for experiment.
Overexpressing of c-Myc
In our study, we used the lentiviral vector pRP[Exp]-EGFP/Puro-EF1A>mMyc to overexpress mouse c-Myc and pRP[Exp]-EGFP/Puro-EF1A > ORF_Stuffer as control, which were constructed and packaged by VectorBuilder. The vector ID are VB231121-1115fks and VB900143-9839ecw respectively, more information about this vector can be found at vectorbuilder.com. Following cell seeding for 24 h, NCTC1469 cells were transfected with 1 μg plasmid respectively using Lipofectamine LTX & PLUS™ Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 48 h transfection, cells were transferred to RPM for experiment.
Statistical analysis
All data are presented as means ± SEM. Statistical analyses were performed in GraphPad Prism 8. The one-way ANOVA was used to compare NG, SMG-A and SMG-S, with Tukey’s post hoc test. For all comparisons, a P value of less than 0.05 was considered significant: *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Freed, L. E. et al. Microgravity cultivation of cells and tissues. Gravitational Space Biol. 12, 57–67 (1999).
He, B., Shi, J., Wang, X., Jiang, H. & Zhu, H. J. Genome-wide pQTL analysis of protein expression regulatory networks in the human liver. BMC Biol. 18, 97 (2020).
Wang, P. et al. Spaceflight/microgravity inhibits the proliferation of hematopoietic stem cells by decreasing Kit-Ras/cAMP-CREB pathway networks as evidenced by RNA-Seq assays. FASEB J. 33, 5903 (2019).
Quynh et al. Simulated microgravity reduces proliferation and reorganizes the cytoskeleton of human umbilical cord mesenchymal stem cells. Physiol. Res. 69, 897 (2020).
Yue, Y. et al. Simulated microgravity altered the proliferation, apoptosis, and extracellular matrix formation of L929 fibroblasts and the transforming growth factor-β1/Smad3 signaling pathway. Skin Res. Technol. 29, e13341 (2023).
Ho, C. N. Q. et al. Simulated microgravity inhibits the proliferation of chang liver cells by attenuation of the major cell cycle regulators and cytoskeletal proteins. Int. J. Mol. Sci. 22, 4550 (2021).
Baba, T. et al. Analysis of gene and protein expression of cytochrome P450 and stress-associated molecules in rat liver after spaceflight. Pathol Int. 58, 589–595 (2008).
Hammond, T. G., Allen, P. L. & Birdsall, H. H. Effects of space flight on mouse liver versus kidney: Gene pathway analyses. Int. J. Mol. Sci. 19, 4106 (2018).
Chen, Y. et al. Upregulation of mir-223 in the rat liver inhibits proliferation of hepatocytes under simulated microgravity. Exp. Mol. Med. 49, e348 (2017).
Beheshti, A. et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 9, 19195 (2019).
Tian, L. Y., Smit, D. J. & Jücker, M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism. Int. J. Mol. Sci. 24, 2652 (2023).
Stengel, S. et al. Suppression of MYC by PI3K/AKT/mTOR pathway inhibition in combination with all-trans retinoic acid treatment for therapeutic gain in acute myeloid leukaemia. Br. J. Haematol. 198, 338–348 (2022).
Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008).
Whitfield, J. R. & Soucek, L. Tumor microenvironment: becoming sick of Myc. Cell. Mol. Life Sci. 69, 931–934 (2012).
Bernard, S. & Eilers, M. Control of cell proliferation and growth by Myc proteins. Results Probl. Cell Differ. 42, 329–342 (2006).
Yu, J. S. L. & Cui, W. Proliferation, survival and metabolism: the role of PI3K/AKT/ mTOR signalling in pluripotency and cell fate determination. Development 143, 3050–3060 (2016).
Skouteris, G. G. & Schröder, C. H. C-myc is required for the G0/G1-S transition of primary hepatocytes stimulated with a deleted form of hepatocyte growth factor. Biochem. J. 316, 879–886 (1996).
Liu, Y. & Wang, E. Transcriptional analysis of normal human fibroblast responses to microgravity stress. Genom. Proteom. Bioinform. 6, 29–41 (2008).
Kumar, A., Tahimic, C. G. T., Almeida, E. A. C. & Globus, R. K. Spaceflight modulates the expression of key oxidative stress and cell cycle related genes in heart. Int. J. Mol. Sci. 22, 9088 (2021).
Xiong, Y. et al. Melatonin ameliorates simulated-microgravity-induced mitochondrial dysfunction and lipid metabolism dysregulation in hepatocytes. FASEB J 37, e23132 (2023).
Bouchard, C. et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18, 5321–5333 (1999).
Almajali, B. et al. Gene expression profiling and protein analysis reveal suppression of the C-Myc oncogene and inhibition JAK/STAT and PI3K/AKT/mTOR signaling by thymoquinone in acute myeloid leukemia cells. Pharmaceuticals 15, 307 (2022).
Zhang, Q., Sakamoto, K. & Wagner, K. U. D-type cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol. Cell Endocrinol. 382, 583–592 (2014).
Fagundes, R. & Teixeira, L. K. Cyclin E/CDK2: DNA replication, replication stress and genomic instability. Front. Cell Dev. Biol. 9, 774845 (2021).
Senoo, H. & Hata, R. Extracellular matrix regulates cell morphology, proliferation, and tissue formation. Kaibogaku zasshi. J. Anatomy. 69, 719–733 (1994).
Dietrichs, D. et al. Three-dimensional growth of prostate cancer cells exposed to simulated microgravity. Front. Cell Dev. Biol. 10, 841017 (2022).
Zhao, H. et al. Effects of simulated microgravity on ultrastructure and apoptosis of choroidal vascular endothelial cells. Front. Physiol. 11, 577325 (2021).
Thompson, E. B. The many roles of c-myc in apoptosis. Ann. Rev. Physiol. 60, 575–600 (1998).
Ciribilli, Y. & Borlak, J. Oncogenomics of c-Myc transgenic mice reveal novel regulators of extracellular signaling, angiogenesis and invasion with clinical significance for human lung adenocarcinoma. Oncotarget 9, 101808 (2018).
Roy, B., Beamon, J., Balint, E. & Reisman, D. Transactivation of the human p53 tumor suppressor gene by c-Myc/Max contributes to elevated mutant p53 expression in some tumors. Mol. Cell Biol. 14, 7805–7815 (1994).
Hsin, I. L., Shen, H. P., Chang, H. Y., Ko, J. L. & Wang, P. H. Suppression of pi3k/akt/mtor/c-myc/mtp53 positive feedback loop induces cell cycle arrest by dual pi3k/mtor inhibitor pqr309 in endometrial cancer cell lines. Cells 10, 2916 (2021).
da Silveira, W. A. et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell 183, 1185–1201.e20 (2020).
Yu, H. et al. STIL promotes tumorigenesis of bladder cancer by activating PI3K/AKT/mTOR signaling pathway and targeting C-Myc. Cancer 14, 5777 (2022).
van Loon, J. J. W. A. Some history and use of the random positioning machine, RPM, in gravity-related research. Adv. Space Res. 39, 1161–1165 (2007).
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2021YFA0719303), the National Natural Science Foundation of China (32271284, 32471282), Guangdong Basic and Applied Basic Research Foundation (2023B1515020109), the Space Application system of China Manned Space Program (YYWT- 0901- EXP-15) and the Sino-European Center of Biomedicine and Health.
Author information
Authors and Affiliations
Contributions
Xiaohua Lei conceived, designed the experiments and manuscript editing. Yue Xiong performed most of the experiments and wrote the manuscript with the help of Ke Zhang and Xiaohua Lei. Chiyuan Ma, Jingtong Xie, and Zekai Chen assisted with the experiments. Ke Zhang designed and supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiong, Y., Ma, C., Xie, J. et al. PI3K/AKT/mTOR pathway and c-Myc inhibit hepatocyte suspension under simulated microgravity. npj Microgravity 11, 88 (2025). https://doi.org/10.1038/s41526-025-00541-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41526-025-00541-5