Abstract
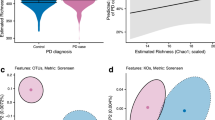
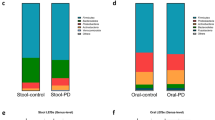
Previous studies suggest there are distinct gut microbial and functional variations in patients with Parkinson’s disease (PwPD) that may reveal potential microbiome signatures or biomarkers to aid in early detection of the disease. In this case-control study, we used whole genome sequencing to compare the stool samples of 55 PwPD to 42 healthy controls (HC) from a public database (BioProject Accession PRJEB39223). For bacterial phyla, we observed a greater relative abundance in Firmicutes and Actinobacteria among PwPD, while that of Bacteroidetes was lower. For phages, PwPD had a greater relative abundance of Siphoviridae, Tectiviridae, and Podoviridae, while Microviridae was lower. Moreover, we described 10 functional pathways that most significantly differed between PwPD and HC (all P < 0.0001). In conclusion, significant differences were observed in gut bacteria, phages, and functional pathways between PwPD and HC that both support and conflict with previous case-control studies and warrant further validation.
Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are not publicly available because the processed outputs incorporate proprietary algorithms and reference databases owned by CosmosID but are available from the corresponding author on reasonable request.
References
Jankovic, J. Parkinson’s disease: clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 79, 368–376 (2008).
Braak, H. & Braak, E. Pathoanatomy of Parkinson’s disease. J. Neurol. 247, II3–II10 (2000).
Cersosimo, M. G. et al. Gastrointestinal manifestations in Parkinson’s disease: prevalence and occurrence before motor symptoms. J. Neurol. 260, 1332–1338 (2013).
Skjærbæk, C., Knudsen, K., Horsager, J. & Borghammer P. Gastrointestinal dysfunction in Parkinson’s Disease. J. Clin. Med. 10, 493 (2010).
Zeng, J., Wang, X., Pan, F. & Mao, Z. The relationship between Parkinson’s disease and gastrointestinal diseases. Front. Aging Neurosci. 14, 955919 (2022).
Adams-Carr, K. L. et al. Constipation preceding Parkinson’s disease: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 87, 710–716 (2016).
Lin, C. H., Lin, J. W., Liu, Y. C., Chang, C. H. & Wu, R. M. Risk of Parkinson’s disease following severe constipation: a nationwide population-based cohort study. Park. Relat. Disord. 20, 1371–1375 (2014).
Lu, S., Jiang, H. Y. & Shi, Y. D. Association between irritable bowel syndrome and Parkinson’s disease: a systematic review and meta-analysis. Acta Neurol. Scand. 145, 442–448 (2022).
Zhang, X. et al. Association between irritable bowel syndrome and risk of parkinson’s disease: a systematic review and meta-analysis. Front. Neurol. 12, 720958 (2021).
Zhu, F. et al. The risk of Parkinson’s disease in inflammatory bowel disease: A systematic review and meta-analysis. Dig. Liver Dis. 51, 38–42 (2019).
Macerollo, A. et al. Colonic diverticular disease: a new risk factor for Parkinson’s disease?. Park. Relat. Disord. 42), 61–65 (2017).
Mulak, A. Bonaz B. Brain-gut-microbiota axis in Parkinson’s disease. World J. Gastroenterol. 21(, 10609–10620 (2015).
Liddle, R. A. Parkinson’s disease from the gut. Brain Res. 1693, 201–206 (2018).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12 (2016).
Sun, M. F. & Shen, Y. Q. Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res. Rev. 45, 53–61 (2018).
Bhattarai, Y. et al. Role of gut microbiota in regulating gastrointestinal dysfunction and motor symptoms in a mouse model of Parkinson’s disease. Gut Microbes 13, 1866974 (2021).
Bedarf, J. R. et al. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 9, 39 (2017).
Chen, S. J. et al. Association of fecal and plasma levels of short-chain fatty acids with gut microbiota and clinical severity in patients with Parkinson disease. Neurology 98, e848–e858 (2022).
Qian, Y. et al. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 143, 2474–2489 (2020).
Nie, S. et al. The link between increased Desulfovibrio and disease severity in Parkinson’s disease. Appl Microbiol. Biotechnol. 107, 3033–3045 (2023).
Qian, Y. et al. Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 70, 194–202 (2018).
Jo, S. et al. Oral and gut dysbiosis leads to functional alterations in Parkinson’s disease. NPJ Parkinsons Dis. 8, 87 (2022).
Yan, Z. et al. Alterations of gut microbiota and metabolome with Parkinson’s disease. Micro. Pathog. 160, 105187 (2021).
Chen, S. J. et al. Plasma metabolites of aromatic amino acids associate with clinical severity and gut microbiota of Parkinson’s disease. NPJ Parkinsons Dis. 9, 165 (2023).
Nishiwaki, H. et al. Meta-analysis of shotgun sequencing of gut microbiota in Parkinson’s disease. NPJ Parkinsons Dis. 10, 106 (2024).
Bolliri, C. et al. Gut microbiota in monozygotic twins discordant for Parkinson’s disease. Ann. Neurol. 92, 631–636 (2022).
De Pablo-Fernandez, E. et al. The faecal metabolome and mycobiome in Parkinson’s disease. Park. Relat. Disord. 95(Feb), 65–69 (2022).
Cirstea, M. S. et al. The gut mycobiome in Parkinson’s Disease. J. Parkinsons Dis. 11, 153–158 (2021).
Metcalfe-Roach, A. et al. Metagenomic analysis reveals large-scale disruptions of the gut microbiome in Parkinson’s disease. Mov. Disord. 39, 1740–1751 (2024).
Wallen, Z. D. et al. Metagenomics of Parkinson’s disease implicates the gut microbiome in multiple disease mechanisms. Nat. Commun. 13, 6958 (2022).
Boktor, J. C. et al. Integrated multi-cohort analysis of the Parkinson’s disease gut metagenome. Mov. Disord. 38, 399–409 (2023).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Mariat, D. et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9, 123 (2009).
Koliada, A. et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 17, 120 (2017).
Magne, F. et al. The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients?. Nutrients 12, 1474 (2020).
Stojanov, S., Berlec, A. & Štrukelj, B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8, 1715 (2020).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Stappenbeck, T. S. et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy 7, 355–374 (2011).
Chen, Z., Li, G. & Liu, J. Autonomic dysfunction in Parkinson’s disease: implications for pathophysiology, diagnosis, and treatment. Neurobiol. Dis. 134, 104700 (2020).
Unger, M. M. et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 32, 66–72 (2016).
Mulak, A. A controversy on the role of short-chain fatty acids in the pathogenesis of Parkinson’s disease. Mov. Disord. 33, 398–401 (2018).
Tan, A. H. et al. Gut microbial ecosystem in Parkinson disease: new clinicobiological insights from multi-omics. Ann. Neurol. 89, 546–559 (2021).
Aho, V. T. E. et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 16, 6 (2021).
Mao, L. et al. Cross-sectional study on the gut microbiome of Parkinson’s disease patients in Central China. Front Microbiol 12, 728479 (2021).
Binda, C. et al. Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 50, 421–428 (2018).
Manrique, P. et al. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA 113, 10400–10405 (2016).
Tetz, G., Brown, S. M., Hao, Y. & Tetz, V. Parkinson’s disease and bacteriophages as its overlooked contributors. Sci. Rep. 8, 10812 (2018).
Tsai, H. H. et al. Hepatitis C virus infection as a risk factor for Parkinson disease: a nationwide cohort study. Neurology 86, 840–846 (2016).
Leta, V. et al. Viruses, parkinsonism and Parkinson’s disease: the past, present and future. J. Neural Transm. 129, 1119–1132 (2022).
Jang, H., Boltz, D. A., Webster, R. G. & Smeyne, R. J. Viral Parkinsonism. Biochim. Biophys. Acta 1792, 714–721 (2009).
Prehn, J. H. M. & Jirström, E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration. Acta Pharm. Sin. 41, 442–446 (2020).
Yu, X. et al. tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 11, 461–469 (2021).
Magee, R., Londin, E. & Rigoutsos, I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Park. Relat. Disord. 65, 203–209 (2019).
Tian, H., Hu, Z. & Wang, C. The therapeutic potential of tRNA-derived small RNAs in neurodegenerative disorders. Aging Dis. 13, 389–401 (2022).
Kuo, M. C., Liu, S. C. H., Hsu, Y. F. & Wu, R. M. The role of noncoding RNAs in Parkinson’s disease: biomarkers and associations with pathogenic pathways. J. Biomed. Sci. 28, 78 (2021).
Paganoni, S. & Schwarzschild, M. A. Urate as a marker of risk and progression of neurodegenerative disease. Neurotherapeutics 14, 148–153 (2017).
Aerqin, Q. et al. Serum uric acid levels in neurodegenerative disorders: a cross-sectional study. J. Alzheimers Dis. 90, 761–773 (2022).
Seifar, F., Dinasarapu, A. R. & Jinnah, H. A. Uric acid in Parkinson’s disease: what is the connection?. Mov. Disord. 37, 2173–2183 (2022).
Dănău, A., Dumitrescu, L., Lefter, A. & Popescu, B. O. Serum uric acid levels in parkinson’s disease: a cross-sectional electronic medical record database study from a tertiary Referral Centre in Romania. Medicina 58, 245 (2022).
Shi, X. et al. Low serum uric acid levels are associated with the nonmotor symptoms and brain gray matter volume in Parkinson’s disease. Neurol. Sci. 43,1747–1754 (2022).
Ari, B. C., Tur, E. K., Domac, F. M. & Kenangil, G. O. Uric acid: the role in the pathophysiology and the prediction in the diagnosis of Parkinson’s disease: a Turkish-based study. Ideggyogy. Sz. 75, 51–59 (2022).
Sartor, B. Intestinal bacterial biofilms modulate mucosal immune responses. J. Immunol. Sci. 2, 13–18 (2018).
Stolzenberg, E. et al. A role for neuronal Alpha-Synuclein in gastrointestinal immunity. J. Innate Immun. 9, 456–463 (2017) .
Christodoulou, D. et al. Reserve flux capacity in the pentose phosphate pathway by NADPH binding is conserved across kingdoms. iScience 19, 1133–1144 (2019).
De Vos, W. M., Tilg, H., Van Hul, M. & Cani, P. D. Gut microbiome and health: mechanistic insights. Gut 71, 1020–1032 (2022).
Hou, J., Xiang, J., Li, D., Liu, X. & Pan, W. Gut microbial response to host metabolic phenotypes. Front. Nutr. 9, 1019430 (2022).
Asnicar, F. et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 27, 321–332 (2021).
Bushnell, B. BBDuk Guide. DOE Joint Genome Institute. Berkeley (CA): U.S. Department of Energy; 2021. Available from: https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbdukguide/.
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Caspi, R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480 (2016).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
The authors would like to thank the patients who participated in the Parkinson Summer School at Bastyr University for providing us with their stool samples. The authors would also like to thank our research assistants, Ali DeMatteo and Fjorda Jusufi, for their help with the study.
Author information
Authors and Affiliations
Contributions
L.K.M., B.E.O., and D.M.W. contributed to the conception of the study and design of the study. B.E.O., D.M.W., and K.M. performed the statistical analysis, data curation, and data visualization. L.K.M. provided supervision and D.J.F. provided management of the project. L.K.M., S.E.E., and J.F. were involved in data collection. S.J.P. and B.E.O. wrote the first draft of the manuscript. All authors: provided critical edits to the manuscript and read and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
B.E.O., D.M.W., and K.M. were employed by CosmosID Inc at the time of the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Park, S.J., Özdinç, B.E., Coker, K.G. et al. Metagenomics indicates an interplay of the microbiome and functional pathways in Parkinson’s disease. npj Parkinsons Dis. (2026). https://doi.org/10.1038/s41531-026-01271-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-026-01271-5