Abstract
Leading gene therapy approaches for Duchenne muscular dystrophy (DMD) using AAV-mediated delivery of microdystrophin (µDys) have shown partial efficacy in patients, contrasting with the favorable outcomes observed in animal models. The identification of effective therapeutic strategies could be accelerated by using human high-throughput DMD models that replicate the molecular complexity driving pathology for accurate screening. To face this challenge, we develop MYOrganoids, an engineered muscle platform derived from patient-induced pluripotent stem cells (iPSC), recapitulating critical hallmarks of DMD, such as fibrosis and muscle dysfunction. We show that co-culture of fibroblasts with iPSC-derived muscle cells during organoid generation is pivotal for functional maturation and muscle force evaluation upon eccentric contractions. Notably, incorporation of DMD fibroblasts induced phenotypic exacerbation in DMD MYOrganoids by unraveling of fibrotic signature and fatiguability through cell-contact and paracrine mechanisms. We then exploited our system to interrogate gene therapy efficacy in this severe context. Although µDys gene transfer improves muscle resistance and partially restores membrane stability, it fails to reduce profibrotic signaling. These findings highlight the persistence of fibrotic activity post-gene therapy in our system, a limitedly explored aspect in DMD models, and provide the opportunity to study mechanisms of dysregulated cellular communication and empower gene therapy efficacy.
Similar content being viewed by others
Introduction
Duchenne muscular dystrophy (DMD; OMIM #310200) is an X-linked disorder that affects one in every 5000 male births1 with no resolutive cure up to date. It is characterized by progressive muscle wasting affecting skeletal muscles primarily and cardiac and respiratory muscles later, thereby causing premature death2. DMD is caused by genetic mutations in the DMD gene, leading to the absence of Dystrophin, an essential protein that provides physical support to myofibers by linking them to the extracellular matrix through the dystrophin glycoprotein complex (DGC)2,3,4,5. The lack of Dystrophin results in a series of muscle membrane breakdowns and repairs, leading to subsequent secondary issues like chronic inflammation and fibrosis6,7,8,9. Fibrosis is an excessive deposition of extracellular matrix components like fibronectin and collagen, triggered by overactivation of transforming growth factor beta (TGF-β) and leading to loss of muscle functionality10,11. Besides being a critical driver of DMD progression, fibrosis also hampers gene therapy efficacy and is therefore paramount to counteract this process that is well-established in patients.
Gene therapy using adeno-associated virus (AAV) is currently the most promising treatment for DMD. Ongoing clinical trials use AAV to deliver short forms of Dystrophin, known as microdystrophin (µDys), which encodes a truncated but functional protein10,11,12,13,14,15,16. However, while the therapeutic effects were unequivocally achieved in DMD animal models17, treated patients expressing robust levels of µDys still show partial therapeutic efficacy and rarely been addressed whether fibrotic activity and signaling were reduced by gene transfer14,18. These observations confirm the limited translatability of results obtained in animal models to human patients. It appears, therefore, crucial to develop time and cost-effective high-throughput models, mimicking the severity of human DMD pathology and its intricate molecular network, suitable for research investigation and therapeutic screening.
In this context, in vitro modeling based on human cells is a valuable option. In particular, the induced pluripotent stem cells (iPSC) technology offers the opportunity to derive an unlimited number of specialized cells from patients for disease modeling and drug screening19,20. Among the in vitro cellular models, organoid-like structures are becoming invaluable for disease modeling as the use of 3D cultures and biomaterials allows the reconstitution of tissue architecture and microenvironment that are instrumental for pathophysiological evaluations21,22. Tissue engineering applications for AAV gene therapy have been exploited mostly in the context of retinopathies20,23,24,25, while only limitedly explored for muscular disorders. Hence, having human DMD models is of utmost importance to advance gene therapy and provide a platform for predictive screening. Although several in vitro 3D systems are accessible for modeling DMD26,27,28,29, their throughput use is limited by the long duration, variability related to the complexity of cellular composition achieved, and lack of disease-specific readouts for muscle function.
Here, we report on the generation of iPSC-derived muscle organoid structure, named hereafter MYOrganoids. We employ and adapt an engineered muscle platform to generate MYOrganoids using a previously reported method for direct iPSC conversion into 2D skeletal muscle cells30,31,32. As a strategy to increase the structural and functional maturation required for pathophysiological studies, we employ for the first time co-culture with fibroblasts, as they are a major source of connective tissue with a recognized regulatory role in muscle differentiation and structure33. Moreover, fibroblasts act as a source of microenvironment cues exerted by their secretory activity, and they are therefore regulators of the muscle niche that undergoes pathological remodeling during disease34,35. Here, we show that fibroblast inclusion leads to improved structural organization and contraction-based functionality of muscle tissue. Moreover, fibroblast integration in a DMD context enables exacerbation of dystrophic traits by direct interaction with muscle cells, unraveling key hallmarks of DMD such as a profibrotic signature and muscle weakness over repeated contractions.
Our study also evaluates for the first time the therapeutic efficacy of AAV-mediated µDys gene transfer in engineering muscle tissues, as proof of concept of their suitability for gene therapy investigation and evaluating potential therapeutics. By using low and high doses of µDys in DMD MYOrganoids, we achieved a dose-dependent response in restoring muscle function, a partial effect on membrane stability, while only a modest reduction of the fibrotic signature in DMD muscles. Our findings indicate that patient-derived MYOrganoids, whose pathogenic traits are exacerbated by a profibrotic environment, effectively recapitulate muscle dysfunction and fibrosis activity. Our model is therefore suitable for the investigation of the fibrotic signaling, orchestrated by muscle and fibroblast populations, as well as the consequences following gene transfer. Our system offers a valuable platform to uncover molecular drivers of dystrophic progression and impaired muscle function, with the potential to enhance gene therapy efficacy and accelerate the discovery of effective treatments for DMD.
Results
Generation of structurally organized 3D human MYOrganoids by direct conversion of iPSC and inclusion of fibroblasts
MYOrganoids were generated from human iPSC committed to differentiating into the myogenic lineage by inducible expression of MyoD and BAF60C30 (referred to as iPSCBM) that can directly generate myotubes. MYOrganoids were prepared starting from iPSCBM after one day from the induction of myogenic genes. The casting procedure was performed through adaptation of an engineered muscle system29,36, which results in the growth of the tissue in a ring format supported by two flexible silicon stretchers. The 3D cultures were kept for 2 days in growth medium, and afterwards medium was replaced for differentiation for another 12 days (Fig. 1A). The differentiation protocol was optimized from the conditions previously reported30,31,32 using myogenic commercial media that ensured the highest myogenic differentiation index in monolayer conditions (Fig. S1A, B). Since cellular heterogeneity, especially of mesenchymal origin, is important for muscle formation29,34,35,37, we included human fibroblasts during the casting procedure to assess whether this would affect muscle organization. For this aim, casting was performed using iPSCBM control cells (CTR1) in the presence or absence of human fibroblasts. Since achieving alignment and differentiation simultaneously necessitates a delicate balance between fibroblasts and muscle cells37, a study was conducted to identify the optimal fibroblast concentration required to achieve correct differentiation and high-level organization. Our analysis revealed that 10% fibroblast inclusion was the optimal condition, as higher concentrations of fibroblasts inhibited differentiation, while lower concentrations were insufficient to promote the alignment of myotubes (Fig. S2A, B). Notably, the optimal 10% fibroblast concentration aligns closely with the proportion of stromal population observed in skeletal muscle, detectable through single-cell and single-nuclei RNA seq analysis38,39,40,41. Fibroblast inclusion accelerated the condensation and growth over time of the muscle rings into a compact structure 8 mm long and 1 mm thick at day 14 (Fig. 1B). The increased compaction observed in fibroblast-containing MYOrganoids was evidenced by their smaller cross-section area (CSA) compared to MYOrganoids lacking fibroblasts (1.8 mm2 vs 0.8 mm2) (Fig. 1C). We then assessed the impact of fibroblast inclusion on muscle structural organization. We assessed fibroblast distribution on longitudinal organoid sections by using Vimentin and Myosin Heavy Chain (MyHC) as fibroblast and muscle markers, respectively. Our analysis revealed that fibroblasts localized both externally and internally the myotubes bundles (42% vs 48%, respectively; Fig. 1D). Given that spatial organization of muscle cells within the ECM plays a key role in their fusion and maturation42, we assessed myotubes alignment and circularity, reflecting the degree of parallel orientation. Myotubes alignment was quantified by measuring the angles between adjacent myotubes43, as delineated by both muscle and membrane markers (MyHC and wheat germ agglutinin-WGA, respectively). The inclusion of fibroblasts resulted in a significant reduction of inter-myotubes angles towards 0 degrees, indicative of aligned orientation, in contrast with MYOrganoids lacking fibroblasts, showing a broader distribution of inter-myotubes angles, an index of poor organization (Fig. 1E, F). These results were also confirmed in another control iPSC line (CTR2) (Fig. S2C). Additionally, circularity of myofibers was assessed on transversal sections stained with WGA, by calculating the ratio between X and Y Feret diameters. MYOrganoids including fibroblasts exhibited significant enhanced myotubes circularity, as indicated by a Feret ratio closer to 1, compared to MYOrganoids without fibroblasts (Fig. 1E, F). These results indicate that fibroblast incorporation during the casting procedure guides skeletal cell orientation, providing structural support for MYOrganoids, which is a prerequisite for maturation.
A Scheme of the protocol used to generate muscle artificial tissues (MYOrganoids) from iPSC (CTR1) committed towards the myogenic lineage by 24 h treatment with doxycycline for inducible expression of Myod and BAF60C transgenes (iPSCBM). The casting procedure included: iPSCBM, fibroblasts when indicated (+/−fibro) and a collagen-based scaffold, within a 48-well plate equipped with silicon pillars. After 2 days in growth medium, the 3D structures were shifted to differentiation medium until day 14 for histological analysis. Pillars image adapted from Santos, JoVE 2021. B Condensation kinetics of MYOrganoids ±fibro. C Quantification of compaction of MYOrganoids with or without fibroblasts, by measurement of organoid cross-section area (CSA). D Fibroblast distribution within MYOrganoids assessed by staining for Vimentin, as a fibroblast maker, and Myosin Heavy Chain (MYHC), as a muscle marker. Numbers indicate percentage of fibroblasts adjacent or external to myotubes (yellow numbers and arrows = external fibro, white numbers and arrows=internal fibro). Scale bar = 100 μm (top panel), 50 μm (lower panel). E Representative longitudinal and cross sections of MYOrganoids ± fibroblasts, immunostained for MYHC and Vimentin. Nuclei were visualized with DAPI. Scale bars 20 µm. Crosses represent X/Y myotubes diameters. F Alignment calculated based on the angle (α) formed between myotubes (α close to 0 corresponds to aligned myotubes, while far from 0 corresponds to not aligned myotubes) by WGA stainings, while circularity from X/Y myotubes diameters ratio (ratio 1 circular, far from 1 not circular). Ratio X/Y diameter is represented by a QQ normality plot. Data were collected from three independent experiments with n = 3. Unpaired two-tailed t test was used (***p < 0.001, ****p < 0.0001).
Increased structural and functional maturation of fibroblast-including MYOrganoids
Since muscle maturation is strictly dependent on the internal myofiber organization, we evaluated the differentiation status of 3D MYOrganoids by analyzing the expression of specific maturation markers, the sarcomere structure and contractile properties. Transversal and longitudinal sections were used to monitor dystrophin (DYS) expression at the sarcolemma (Fig. 2A) and sarcomeric α-actinin (SAA) localization for assessment of the striation pattern typical of mature myotubes (Fig. 2B). Dystrophin was properly localized to the sarcolemma of myotubes derived from MYOrganoids, including fibroblasts, and was significantly more expressed than MYOrganoids without fibroblasts at both protein and mRNA levels (Figs. 2D and S2D). Increased maturation of myotubes within MYOrganoids including fibroblasts was supported by the analysis of fusion and striation indices, using muscle and membrane markers to delineate myotube structures (SAA and WGA, respectively) (Fig. 2B). Quantitative analysis of immunofluorescence shows a significantly higher fusion index, with 20% of myotubes including more than 3 nuclei (Fig. 2E), as well as higher maturation index, evidenced by an increase proportion of striated myotubes characterized by clear spaced sarcomeric bands (~60% in + Fibro vs 20% in −Fibro condition) (Fig. 2F). In contrast, MYOrganoids lacking fibroblasts exhibited myotubes that were smaller, disorganized and displayed minimal or poorly defined striation pattern (Fig. 2F). These findings highlight the contribution of fibroblasts in promoting myotubes maturation through fusion and multinucleation. The proper sarcomeric organization was also confirmed by electron microscopy, where we could detect longer, properly formed Z patterning and the presence of I and A bands along with an overall increase of sarcomeric density and alignment (Fig. 2C). Consistently, MYOrganoids containing fibroblasts showed wider Z-line (Fig. 2G), an index of higher maturation of sarcomeres44. We further performed gene expression analysis for terminal differentiation markers such as muscle creatine kinase (MCK) and myosin heavy chain (MYH) isoforms that are present in the adult muscle, such as MYH2 and MYH7, representative of fast-twitch (Type II) or slow-twitch adult fiber type (Type I), respectively. Higher expression of these genes in MYOrganoids containing fibroblasts confirmed the acquisition of a more mature state, compared to MYOrganoids without fibroblasts (Fig. 2H).
A Representative transversal sections of MYOrganoids ± fibroblasts stained for Dystrophin (DYS). Scale bar: 40 µm. Nuclei were visualized with DAPI. B Representative longitudinal sections of MYOrganoids ± fibroblasts (fibro), immunostained for Sarcomeric α-Actinin (SAA) and nuclei visualized with DAPI. Scale bars: 40 µm, enlargement 20 µm. C Transmission electron microscopy images showing sarcomeric structures. Orange arrows: Z-lines. Scale bar: 500 nm. D DYSTROPHIN staining quantification represented as mean intensity fluorescence. N = 5 E Quantification of fusion index calculated as % Myosin Heavy Chain positive (MYHC+) myotubes containing different numbers of nuclei (n) and delimited by WGA staining at the membrane. N = 3. F Quantification of striation index as a percentage of regularly spaced bands-presenting myotubes of the total number of myofibers delineated by wheat germ agglutinin (WGA). N = 4 G Width of Z-line visualized by Electron microscopy, in MYOrganoids with and without inclusion of fibroblasts, obtained from four fields in each biological replicate. N = 4. H Gene expression analysis of MCK, MYH2 and MYH7, reported as gene expression relative to MYOD-expressing population. N = 3–4 I Representative transversal section of MYOrganoids + fibroblasts stained for SAA, MYH IIb and MYH I, and relative quantification obtained from eight fields in each condition. Scale bar = 30 µm, N = 2. The percentage of MYH fluorescence was calculated as the MFI of each MYH over the total fluorescence in all the channels. J Contractile muscle force analysis of MYOrganoids using a muscle-strip-based organ-bath system. Lo, the optimal length is used for normalization of force data (see methods). K Force-frequency plot for MYOrganoid with and without fibroblasts. L Representative tetanic traces of MYOrganoids with and without the inclusion of fibroblasts, stimulated at 125 Hz, for 2 seconds. M Normalized tetanic force peak in MYOrganoids ± fibroblasts. N = 9). Data are presented as mean ± SEM. Unpaired t test was applied for statistical analysis in D–H and M (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001), while in G, MYH% quantification was analyzed using one-way analysis of variance with Tukey’s multiple comparison tests (*p < 0.05).
Interestingly, although our MYOrganoids still express embryonic myosin (such as MYH3, Fig. S2E)—a feature consistent with the molecular profile of iPSC-derived muscles undergoing maturation45,46—histological analysis revealed a heterogeneous distribution of muscle fiber type. Specifically, we identified predominance of fast-twitch fibers, with Type IIb fibers (60%) and Type IIa (20%), and a minority of slow-twitch fibers, Type I (20%), confirming prevalence of fatigue-prone myotubes, relevant to the objectives of our study (Fig. 2I).
We then assessed whether our MYOrganoids were functional by evaluating their contractile properties in response to electrical stimulation, using a muscle organ-bath system based on electrical pacing47. For force measurement, MYOrganoids were transferred to muscle strip chambers and stretched until reaching the optimal length (Lo) for functional analysis (Fig. 2J). The force-frequency relationship was established to identify the electric pulse frequency at which MYOrganoids developed maximal force at Lo (Fig. 2K). Isometric force analysis, conducted with a tetanus (125 Hz for 2-second trains), revealed higher tetanic force in MYOrganoids containing fibroblasts compared to the control (Fig. 2L). Tetanic peak values were then normalized for the cross-sectional area (CSA) using the weight and the Lo of each MYOrganoid, as previously described48,49, and expressed as specific tetanic force (mN/mm2). MYOrganoid containing fibroblasts exhibited an average peak of specific tetanic force of 0.6 mN/mm2 compared to 0.3 mN/mm2 in MYOrganoids lacking fibroblasts (Fig. 2M). These results indicate that the inclusion of fibroblasts enhances both the structural organization and functional maturation of MYOrganoids, thereby enabling robust assessment of contractile force in response to electrical pacing.
Inclusion of fibroblasts in DMD iPSC-derived MYOrganoids leads to increased muscle force loss and profibrotic signature
The improved muscle organization and functional maturation shown by MYOrganoids, including fibroblasts, prompted us to exploit fibroblast features in disease modeling for DMD, where their role in disease progression is well known50,51. Specifically, we incorporated DMD fibroblasts to recapitulate the pathogenic microenvironment arising from their profibrotic activity through tissue remodeling and matrix deposition51,52. For that purpose, we used three DMD iPSC lines with different DMD mutations, a deletion of exon 45 (DMDdEx45), a deletion of exons 8-43 (DMDdEx8-43) and a deletion of exons 8-9 (DMDdEx8-9) with their isogenic control, obtained by CRISPR-CAS9 correction to restore dystrophin expression53 (hereafter called IsoCTR). We additionally used two control iPSC lines derived from healthy patients (CTR1, CTR2). Control and DMD MYOrganoids were generated from control and DMD iPSC with healthy or DMD human immortalized fibroblasts for functional and transcriptomic analysis (Fig. 3A). The myogenic differentiation ability of both CTR and DMD iPSC lines was first evaluated in 2D cultures by immunofluorescence, which demonstrates that all the cell lines used in our study showed comparable and high myogenic potential (Fig. S3A). Histological characterization in MYOrganoids confirmed the absence of dystrophin protein in DMD MYOrganoids and efficient myogenic differentiation and maturation, both in CTR and DMD MYOrganoids, revealed by the striation pattern visualized by SAA-stained sections (Fig. 3B). Furthermore, gene expression analysis of MYOrganoids derived from CTR iPSC, both with and without fibroblasts, revealed higher expression of DMD and MCK in the presence of fibroblasts (Fig. S3B). Similarly, MYOrganoids generated from DMD iPSCs exhibited increased MCK when fibroblasts were included (Fig. S3C), confirming the positive role of fibroblasts in enhancing maturation also in a DMD context.
A Overview of MYOrganoids generation from control iPSC (CTR1 and CTR2), DMD iPSC (DMDdEx45, DMDdEx8-43) and isogenic iPSC (IsoCTR and DMDdEx8-9) including control or dystrophic fibroblasts respectively for histological characterization, force and transcriptomic analysis. B Immunostaining of MYOrganoids cross sections for Dystrophin (Dys) on transversal sections, Sarcomeric a-actinin (SAA) on longitudinal sections, and DAPI for nuclei. Scale bar: 30 µm (C) Evans blue dye (EBD) fluorescence in IsoCTR (orange) and DMDdEx8-9 (blue) MYOrganoids ± fibroblasts (fibro) before and after damage induction with Notexin (NTX) or 1 eccentric contraction (1×) EBD fluorescence was calculated as a percentage of total MYHC + cells. D Muscle force loss calculated as % of drop force between two isometric contractions (ISO) performed before and after 10 repetitions of ECC in all CTR and all DMD MYOrganoids ± respective fibroblasts. N = 6. E Half relaxation time (Time ½ Fmax) in CTR and DMD MYOrganoids ± fibroblasts. N = 4–6. F Gene Ontology enrichment analysis (GO TERM) from RNA-seq data in isogenic iPSC-derived CTR (IsoCTR) and DMD (DMDdEx8-9). The scale of color is representative of the p adjusted (p.adj) N = 4, biological replicates. G Clustered heatmap of fibrotic Differentially Expressed Genes (DEG) relative to DMD vs IsoCTR (left). Color scale represents the Z-score (red is higher, blue lower). H Relative gene expression of Fibronectin (FN1), Collagen (COL1A1) and Periostin (POSTN) in all iPSC CTR and DMD-derived MYOrganoids with or without fibroblasts. For FN1, N = 4 and for COL1A1 N = 3. I Capillary western blot for pSMAD2/3 (60 kDa) from all CTR and DMD MYOrganoids protein lysate and relative quantification. J Representative images of immunostaining on longitudinal sections showing activation of TGF-β signaling by immunostaining of MYHC (magenta), GFP (green) and pSMAD (red). Nuclei are visualized with DAPI, and nuclei belonging to myotubes are circled in white, while those belonging to fibroblasts are in yellow. Graph on the right is the relative quantification of pSMAD+ cells in myotubes. Scale bar: 25 µm, N = 3. For all panel, data are presented as mean ± SEM. D–E/H unpaired two-tailed t test was performed for statistical purposes. J a two-way analysis of variance with Šidák correction test was performed (F = 1,12, degree of freedom = 12) (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
To assess whether DMD MYOrganoids display hallmarks of DMD pathophysiology, we evaluated muscle function, which represents one of the most difficult challenges in establishing therapeutic readouts with in vitro systems. Tetanic force analysis after just isometric contractions led to significant differences between Control and DMD iPSC MYOrganoids in the isogenic cell lines, while no differences were observed between the other control and DMD groups (Fig. S4A). Therefore, to identify reliable force parameters reflecting the defective DMD muscle performance common in all patients, iPSC-derived MYOrganoids were subjected to eccentric contractions (ECC), which play a critical role in the disease progression of DMD54 by triggering muscle fiber damage and sarcomere disruption14. To evaluate ECC-induced sarcolemmal damage, we performed Evans Blue Dye (EBD) incorporation experiments in MYOrganoids without contraction and with 1X ECC contraction versus Notexin (NTX)-induced damaged organoids, as a positive control. Histological staining and relative quantification revealed that EBD incorporation following 1X ECC repetition in DMD organoids, including fibroblasts, was higher compared to 1X ECC CTR organoids and to unstimulated DMD organoids and shows that the first ECC starts eliciting a sarcolemma damage that involves 12% of DMD myotubes (Figs. 3C and S4B). Interestingly, EBD incorporation after NTX-induced damage was higher in DMD MYOrganoids with fibroblasts compared to the CTR condition (Fig. 3C), confirming the higher sensitivity of DMD sarcolemma to ECC-induced damage. Therefore, we used ECC repetitions, with 1-minute rest in between, as an induced muscle fatigue protocol, similar to what is being used in patients55. DMD MYOrganoids incorporating DMD fibroblasts demonstrated a significant drop in contractile force when compared to CTR organoids starting from the 4th eccentric repetition (Fig. S4C). Importantly, Myorganoids recovered most of their force capacity after 60 minutes, in agreement with what was observed after ECC-protocol in mdx model56 (Fig. S4D), supporting reproducibility of the muscle mechanics with our system. We then assessed muscle force loss, resulting from either fatigue or transient muscle damage, by measuring the decline in force between isometric contractions performed before and after the eccentric repetition protocol. The analysis showed a significantly higher fatigue index in DMD MYOrganoids as compared to CTR MYOrganoids and remarkably, this phenomenon was highly accentuated by the presence of fibroblasts (Fig. 3D). To check whether the impact on disease exacerbation and increase of difference in fatiguability between CTR and DMD MYOrganoids was affected by fibroblast genetic source, we employed three additional fibroblast cell lines from either healthy and DMD individuals to generate CTR and DMD MYOrganoids. Muscle force analysis confirmed that the fatigue index in DMD MYOrganoids was significantly higher than CTR MYOrganoids, and that this exacerbation effect was recapitulated using control or DMD fibroblasts from different sources, indicating an effect due to the fibroblast activity and not influenced by the genetic background (Fig. S4E).
Finally, we assessed the half relaxation time (Time ½ Fmax), a relevant parameter for evaluating muscle function in DMD, as prolonged relaxation time is a feature of fatigued muscle and indicative of an impaired calcium reuptake following contraction57,58. Our results demonstrate that DMD MYOrganoids exhibited altered calcium kinetics exclusively in the presence DMD fibroblasts, as shown by a significantly higher time required for muscle force to decline to 50% of its peak value post-contraction (Fig. 3E). Collectively, these data indicate that DMD MYOrganoids including DMD fibroblasts, exacerbate loss of muscle resistance and increase in fatiguability. The finding also demonstrates that muscle force deficit evaluated by eccentric-based fatigue protocol, and analysis of half relaxation time are meaningful and reliable therapeutic readouts, as significant differences were detected across all CTR and DMD MYOrganoids groups including fibroblasts.
We then performed transcriptomic analysis in the isogenic iPSC lines-derived MYOrganoids after 24 h from ECC protocol, to better characterize the pathogenic hallmarks introduced by fibroblast incorporation within the organoids in pathological conditions. Analysis of significant (p.adj < 0.05) and relevantly different transcripts (abs log2FoldChange > 1) revealed 4610 differentially expressed genes (DEGs) between DMDdEx8-9 and IsoCTR MYOrganoids, of which 2181 upregulated and 2429 downregulated (Fig. S5A, B). Of those, we identified genes involved in decreased muscular contraction (e.g. muscle system process, muscle contraction, sarcoplasmic reticulum calcium ion transport and actin-myosin filament sliding) as well as reduced energetic molecular process (e.g. oxidative phosphorylation, mitochondrial respiratory chain complex assembly, ATP synthesis coupled electron transport), as depicted by gene ontology biological process (GOBP) enrichment analysis (Fig. 3F, Table S1). Remarkably, among the upregulated pathways in DMD MYOrganoids, we identified higher collagen fibril organization with an enrichment score of +0.6 (Fig. 3F). This GO analysis was also confirmed by Kyoto encyclopedia of genes and genome (KEGG) enrichment analysis, which showed a downregulation of oxidative phosphorylation and an upregulation of ECM receptor interaction (Fig. S3C). Interestingly, the analysis of genes involved in the ECM remodeling revealed an upregulation of genes coding for collagens, laminins, metalloproteases, integrins and TGF-β-related genes such as TGF-β receptor (TGFBR1) and the ncRNA inhibitor of TGF-β (TGFB2-AS1) in DMD MYOrganoids compared to isogenic control (Fig. 3G, Table S2). Additionally, we found increased levels of latent TGF-β binding proteins (LTBP 2) in dystrophic MYOrganoids, indicating that TGF-β is activated in the DMD context and contributes to ECM remodeling (Table S2). A few ECM-related proteins (such as LAMA2, MMP1) show opposite trends, which can occur in muscle actively remodeling, like in our post-exercise conditions.
The increased profibrotic signature in DMD iPSC-derived MYOrganoids was then confirmed in all CTR and DMD iPSC-derived MYOrganoids, by evaluation of the expression of key fibrotic genes, such as Fibronectin-1 (FN1), Collagen-1 (COL1A1), Periostin (POSTN) (Fig. 3H). We additionally checked by qRT-PCR, Fibroblast Activation Protein (FAP), and Extra Domain A splice variant of fibronectin 1 (EDA-FN1), which are markers specifically associated with pathological fibrosis and used as a clinical biomarker of disease progression59 (Fig. S5D, E). Importantly, the differences in the expression of fibrotic markers across all CTR and DMD iPSC-derived MYOrganoids were significant only when including fibroblasts (Fig. 3H and Fig. S5D, E). We then evaluated the overall increase of profibrotic activity in all DMD iPSC-derived MYOrganoids, by looking at the protein expression level of phosphorylated SMAD3 (pSMAD3), the transcriptional effector of the canonical TGFβ pathway. Capillary western blot quantification showed significantly higher expression levels of pSMAD3 across all DMD MYOrganoids (Fig. 3I). We further examined at the histological level the presence of activated TGF-β signaling in DMD organoids in the muscle population, identified by MYHC-positive cells, and in fibroblasts stably expressing GFP. Notably, DMDdEx8-9-derived MYOrganoids showed increased pSMAD3-positive nuclei both in myotubes (37%) and fibroblasts (25%) (Fig. 3J). Overall, these data demonstrate that fibroblast integration into muscle organoids enables unraveling of fibrotic signature and muscle dysfunction across multiple iPSC lines, thereby establishing reliable readouts for therapeutic screening.
Cell-contact and paracrine mechanisms drive profibrotic signatures in DMD MYOrganoids
We then sought to gain insights into the contribution of fibroblasts in the recapitulation of key pathogenic hallmarks in DMD MYOrganoids. Given that fibroblasts are known to primarily act through profibrotic secreted molecules60,61,62 we analyzed the secretome of both control (CTR) and DMD MYOrganoids (MyoT + Fibro), by looking at key secreted factors by ELISA assay, and compared it to that of single-cell 3D cultures (Fibro-only and MyoT-only) (Fig. 4A). Analysis of culture media on day 14 showed elevated levels of secreted TGF-β, Collagen 4 (COL4) and Fibronectin 1 (FN1) in Fibro-only 3D cultures and DMD MYOrganoids co-cultured with fibroblasts. No significant differences in TGF-β and FN1 secretion were observed between control and DMD in Fibro-only and MyoT-only 3D cultures (Fig. 4A), while higher secretion of COL4 was observed in DMD Fibro-only vs control. Gene expression analysis paralleled secretome data, except for DMD MyoT-only- culture expressing more TGFβ and FN1, consistent with the constitutive TGFβ activation observed in DMD muscle cells30 (Fig. S6A). These data indicate that DMD MYOrganoids, where muscle and fibroblasts are co-cultured, exhibit an increased secretion and expression of fibrotic factors.
A TGF-β, Collagen 4 (COL4) and Fibronectin 1 (FN1) ELISA on culture media collected at day 14 in vitro from fibroblasts (Fibro-only), myotubes (MyoT) and co-culture of both (MyoT + Fibro = MYOrganoids) from IsoCTR or DMDdEx8-9 cells. N = 4 B Scheme of conditioned medium (CM) exchange. DMD organoid CM was collected from day in vitro 7 and used to treat every other day CTR MyoT-only 3D cultures. Pillars image adapted from Santos, JoVE 2021 (C) TGF-β, FN1 and COL4 ELISA performed on media collected from CTR MyoT-only (top panels) and CTR Fibro-only (lower panels) 3D cultures treated or not with DMD-organoid CM. N = 4 D TGF-β, FN1 and COL4 secreted proteins in culture media collected from CTR organoid (MyoT + Fibro) treated with DMD-organoid CM vs untreated. N = 4 (E) TGF-β, COL1A1 and FN1 gene expression in CTR organoid (MyoT+ Fibro) treated or not with DMD-organoid CM. N = 5 F Secreted TGF-β, FN1 and COL4 detected in culture media at day in vitro 14 in CTR organoid (CTR MyoT + CTR Fibro), CTR mixed organoid (CTR MyoT + DMD Fibro), DMD organoid (DMD MyoT + DMD Fibro) and DMD mixed organoid (DMD MyoT + CTR Fibro). N = 3–4. G Muscle force loss indicated as percentage of drop force (Iso2-Iso1) in organoid (CTR MyoT + CTR Fibro), CTR mixed organoid (CTR MyoT + DMD Fibro), DMD organoid (DMD MyoT + DMD Fibro) and DMD mixed organoid (DMD MyoT + CTR Fibro). N = 6. For all panels, data are represented as average ± SEM. A, C, D, F measurements of secreted proteins were normalized against the total protein present in the collected media. A–E Unpaired Student t test was applied for statistical significance. E–G one-way analysis of variance and multiple comparison with Tukey’s correction of each condition vs CTR and DMD organoid was performed DMD vs CTR. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns = not significant).
To determine the origin of paracrine signaling, we evaluated the impact of conditioned media (CM) derived from either DMD Fibro-only, MyoT-only and DMD fibro/MyoT co-culture (MYOrganoids) on the single cultures. CM was collected from day 7 of in vitro growth every 48 h until day 14, then applied to the 3D cultures for the same period and assessed expression and secretion of fibrotic factors along with muscle force analysis. No differences were observed in the secretion and gene expression of TGF-β, FN1 and COL4 in MyoT-only or Fibro-only cultures treated with the complementary CM, indicating a lack of reciprocal paracrine influence between fibroblasts and myotubes from the single cultures (Fig. S6B–E). By contrast, CM derived from DMD MYOrganoids, previously shown to be enriched in profibrotic factors (Fig. 4A), induced a significant increase in the secretion of TGF-β, FN1 and COL4 in the CTR MyoT-only 3D cultures, but not in the CTR Fibro-only 3D cultures (Fig. 4B, C). We then evaluated the ability of DMD-organoid CM to induce a profibrotic signature on CTR organoids. We observed an increased expression and secretion of TGF-β, FN1 and COL4 (Fig. 4D, E). These results suggest that DMD organoid-CM induces profibrotic signaling in CTR organoids by acting primarily on CTR myotubes.
The lack of responsiveness to DMD MYOrganoids CM of the fibroblast population (Fig. 4C) suggested that cell-contact mechanisms may participate in inducing pathogenic modeling in DMD MYOrganoids. To address this question, we created mixed organoids combining fibroblasts and myotubes from control and dystrophic genotypes (CTR fibroblasts with DMD myotubes, and DMD fibroblasts with CTR myotubes) to determine whether direct cell–cell contact between fibroblasts and myotubes of different genotypes was required for the induction of profibrotic signaling. No significant differences were observed in the secretion or expression of TGF-β in the mixed organoids compared to classic MYOrganoids, whereas FN1 and COL4 secretion increased in the mixed organoids combining CTR myotubes with DMD fibroblasts compared to CTR organoids (Figs. 4F and S6F). Additionally, organoids containing DMD MyoT and CTR fibroblasts showed a decreased secretion of COL4 compared to DMD organoids (Fig. 4F). Subsequent analysis of expression of profibrotic genes revealed a marked upregulation of COL1A1 and FN1 expression in organoids containing DMD fibroblasts (CTR M + DMD F), indicating a master role of DMD fibroblasts in driving profibrotic environment through direct cell contact with myotubes (Fig. S6F). Conversely, organoids composed of CTR fibroblasts (DMD M + WT F) exhibited reduced COL1A1 and FN1 expression (Fig. S6F). Force analysis of the mixed organoids revealed a stronger force deficit versus CTR organoids, when at least one DMD population is included in the mixed organoids, which is, however, statistically significant from organoids composed of both DMD populations (Fig. 4G). These data suggest that, while DMD myotubes or fibroblasts can partially induce fibrotic and dystrophic changes in muscle organoids by reciprocal signaling, the robust dystrophic phenotype only emerges when DMD fibroblasts and DMD muscle cells are co-cultured, indicating that both synergistic paracrine influence and direct cellular interaction between these cell types are essential to fully manifest the DMD phenotype.
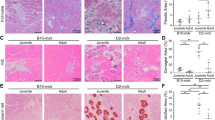
AAV-µDys gene transfer rescues muscle resistance while partially restoring DGC components in DMD MYOrganoids
As proof of concept that MYOrganoids were suitable as a screening platform for gene therapy, we performed AAV-mediated delivery of µDys and assessed its therapeutic efficacy in the DMD context. We used AAV9 capsid for this aim, as this viral serotype was used in recent clinical trials and showed a high transduction rate in patients’ myofibers11,63,64,65. We infected the organoids with a codon-optimized µDys gene (dR4-23, same protein product used in clinical trials14) under the control of the muscle-specific spc512 promoter13 for gene transfer using AAV9 (AAV9-µDys) in all DMD MYOrganoids iPSC (dEx45, dEx8-43 and dEx8-9 with isogenic control, IsoCTR) and assessed gene transfer efficiency, muscle force analysis and membrane stability assessment (Fig. 5A). We first optimized the infection conditions using the reporter AAV9-CMV-GFP in CTR MYOrganoids, including fibroblasts. Infection was performed on day 7 of the differentiation protocol, diluting AAV particles directly in the medium, and maintained for an additional 7 days (Fig. S7A–C). Optimal low and high AAV9-µDys doses were established based on previous dosing studies to have intermediate transduction levels at low doses (1E + 09 vg/MYOrganoid), and high transduction levels at high doses (5E + 10 vg/MYOrganoid) (Fig. S7D). Gene transfer efficiency was evaluated by quantification of viral copy number (VCN) on genomic DNA and by the expression level of the transgene and the encoded protein, showing a clear dose-dependent entry and expression of µDys in DMD MYOrganoids (Fig. 5B–D, S8A). As expected by using a muscle-specific promoter, only the muscle cells (MyoT) but not the fibroblasts expressed the transgene after infection of AAV9-uDys in the single populations (Fig. S8B). To confirm that the observed effects were specifically due to µDys expression rather than AAV infection alone, we infected DMD MYOrganoids with a control vector containing a dystrophin fragment lacking a promoter (hereafter called empty vector), which infection did not result in protein expression (Fig. S8C–E). We then tested whether µDys gene transfer could rescue muscle performance in DMD MYOrganoids. To this aim, infected DMD MYOrganoids were subjected to the eccentric exercise protocol, and muscle force loss and relaxation time was calculated as previously described (Fig. 3). Our analysis shows a dose-dependent attenuation of muscle force loss in DMD MYOrganoids after infection, reaching significance only for DMDdEx45 and DMDdEx8-9 MYOrganoids treated at high doses of AAV9-µDys, as compared to MYOrganoids infected with the empty vector that did not show any differences in the force index and behaved as untreated DMD organoids (Fig. 5E). Analysis of half relaxation time (T ½ Fmax) show a decrease across all cell lines, supporting the positive effect of µDys in muscle function improvement (Fig. 5F). Overall, the muscle force data confirmed that ectopic µDys expression rescues muscle function, although at different extent across patients-derived cell lines.
A Scheme of AAV9-µDys infection and doses used in DMD iPSC-derived MYOrganoids. B Evaluation of gene transfer efficiency by viral copy number (VCN) analysis. C µDys gene expression evaluated by RT-ddPCR represented as fold change to GAPDH gene expression. D Protein expression analysis by capillary western blot in isogenic iPSC (IsoCTR and DMDdEx8-9) derived MYOrganoids infected with low and high doses of µDys versus non-infected (–) IsoCTR (N = 3). E Percentage of drop force to calculate percentage of muscle force loss (Iso2-Iso1) after 10 eccentric contractions in isogenic cell lines, IsoCTR and DMD dEx8-9, and CTR1, CTR2 and DMD dEx45, DMDdEx8-43-derived MYOrganoids receiving low and high doses of µDys. N = 6 (F) Half decreasing time (Time ½ Fmax) in all CTR and DMD MYOrganoids treated or not with µDys at low and high dose. N = 5–8. G Histological evaluation of Dystrophin Glycoprotein Complex (DGC) upon infection, by immunostaining for Dystrophin (DYS), α-Dystroglycan (α-DG) and β- Dystroglycan (β-DG) in transversal sections of Isogenic CTR and DMD-iPSC-derived MYOrganoids. Scale bar: 40 µm and H relative quantification in all control and iPSC-derived MYOrganoids N = 4–5. Data are presented as means ± SEM. Statistical analysis was performed with an two-way analysis of variance test (for D) and ordinary one-way analysis of variance test (for all the other panels) with Šidák correction in multiple comparison tests(*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant) with multiple comparisons corrected with Dunnett’s test (F = 3,24 and degree of freedom = 24).
Because Dystrophin exerts its biomechanical support by holding the DGC at the membrane, we monitored key components of this complex, such as the transmembrane β-Dystroglycan (β-DG), directly binding Dystrophin, and the extracellular α-Dystroglycan (α-DG), whose proper expression and localization are impaired in the absence of dystrophin66. To this aim, DMD MYOrganoids infected with low and high doses of µDys were subjected to histological analysis. Quantification of µDys showed around 25–30% of dystrophin-positive myotubes at low dose compared to 85% in the high dose condition in DMD dEx8-9 (Fig. 5G, H) and dEx45 and 65% in the DMD dEx8-43 MYOrganoids (Fig. S9A, B). Immunostaining on transversal MYOrganoids showed a dose-dependent yet not complete restoration of β-DG in all DMD iPSC-derived MYOrganoids. Interestingly, even high doses ensuring nearly total Dystrophin transduction in all the DMD MYOrganoids did not significantly restore α-DG (~10% in DMDdEx8-9 and DMDdEx8-43 and ~30% in DMDdEx45) (Fig. 5G, H). Importantly, these data demonstrate a clear improvement in muscle strength after µDys gene transfer, but only a partial restoration of Dystrophin-associated components.
Optimal µDys gene transfer partially corrects the transcriptomic profile of DMD MYOrganoids and does not rescue the profibrotic signature
To examine the effect of µDys on the signaling pathways and molecular processes characteristics of dystrophic pathology, we performed bulk RNA-seq analysis comparing the corrected DMD dEx8-9 (IsoCTR) and DMDdEx8-9 MYOrganoids untreated and treated with a high dose of AAV9-µDys (n = 4).
Principal component analysis (PCA) of the top 10,000 genes with the highest variance showed a distinct transcriptomic profile of DMD organoids treated with µDys (dEx8-9 + µDys high) and untreated (dEx8-9) compared to isogenic controls (IsoCTR) (Fig. S10A). To evaluate the effect of µDys in restoring key pathways involved in DMD, we first compared the transcriptomic profile of µDys-treated DMD organoids to untreated DMD organoids (dEx8-9 µDys vs dEx8-9), and then to the isogenic control (dEx8-9 µDys vs IsoCTR). Particularly, µDys-treated DMD organoids showed a total of 327 DEGs, of which 154 upregulated and 173 downregulated, compared to untreated organoids (Fig. 6A, Table S3). Of those 327 DEGs, 52% (170/327) are common with the DEGs that were found dysregulated in DMD organoids when compared to the isogenic control (Fig. S10B). Therefore, we performed the gene ontology enrichment analysis of the common category to see the effect of μDys on the DMD-dysregulated pathways. This analysis, where gene sets are ordered by normalized enrichment score (NES) revealed increased expression of genes involved in the microtubule organization, (GOBP: Microtubule organizing center organization NES = +2.31), cytoskeleton organization (GOBP: Protein localization to cytoskeleton NES = + 1.99), and a decreased expression of genes involved in the inflammation (GOBP: Regulation of inflammatory response, NES = −1.45) (Fig. 6B).
A, C Volcano plot showing the differential gene expression in treated (dEx8-9-µDys) versus untreated (dEx8-9) DMD MYOrganoids (A) or DMD dEx8-9-µDys vs isogenic control (IsoCTR (C). Upregulated genes are shown in red, while downregulated genes are shown in blue. Numbers indicate the amount of downregulated and upregulated genes. B, D Gene ontology enrichment analysis of treated (dEx8-9-µDys) versus untreated (dEx8-9) DMD MYOrganoids (B) or dEx8-9-µDys vs IsoCTR (D). Normalized enrichment score (NES) and adjusted p value (p.adj) are indicated for each subcategory. X axis indicates the running enrichment score, while the y axis indicates the position in the ranked gene list. E Clustered Heat map and enlargement depicting relative transcript levels of differentially expressed genes in corrected DMD (IsoCTR) and DMD MYOrganoids transcriptomics treated or not with µDystrophin (dEx8-9 and dEx8-9 µDys high). Lower and higher expressions are depicted in blue and red, respectively. The data points above the significance threshold (p adjusted < 0.05, fold2change > 2) are marked in blue (downregulated) and red (upregulated), and others are marked in gray (not significant). F Relative gene expression of TGF-β, COL1A1, FN1 and ITGA10 in Isogenic control (IsoCTR), DMD dEx8-9 untreated and treated with empty vector and μDys at low and high dose. G Representative pictures of histological staining on longitudinal sections for phospho-SMAD (pSMAD, red), myosin heavy chain (MYHC, magenta marking the myotubes), GFP (green, marking the fibroblasts) and nuclei with DAPI in IsoCTR and µDys-treated DMD dEx8-9 MYOrganoids, and relative quantification of pSMAD3-positive nuclei in percentage in all CTR and DMD MYOrganoids. Myotube’s nuclei are circled in white, while fibroblasts’s nuclei are indicated in yellow. Scale bar = 20 µm. F, G data are presented as means ± SEM. F statistical analysis was performed with an ordinary one-way analysis of variance test with Tukey multiple comparison test (*p < 0.05, **p < 0.01, ns = not significant), while for G two-way analysis of variance test with multiple comparison and Šidák correction was performed (*p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001 ns = not significant).
We then analyzed the DEGs between the treated organoids and the isogenic control to check the transcriptomic signature not restored by µDys (dEx8-9-µDys vs IsoCTR). Particularly, µDys-treated organoids present 7818 DEGs (4004 upregulated and 3814 downregulated) when compared to the corrected isogenic organoids (Fig. 6C). Gene ontology analysis of the DEGs between µDys-treated DMD organoids and IsoCTR organoids revealed the pathways that are not restored by the gene transfer. Particularly, treated organoids showed persistent downregulation of genes involved in calcium transport (GOBP: Sarcoplasmic reticulum calcium ion transport, NES = -2.15), oxidative phosphorylation (GOBP: Oxidative phosphorylation, NES = −2.29) and upregulation of TGFβ signaling (Hallmark TGFβ signaling, NES = +1.57), when compared to isogenic controls (Fig. 6D, Table S4). Consistently, a clustered heatmap of all 1716 most significant differentially expressed genes (DEGs) further highlighted the transcriptomic differences between control and DMD organoids (Fig. 6E).
Analysis of Z-score of fibrotic genes among the three conditions (IsoCTR, DMDdEx8-9 and DMDdEx8-9 treated with µDys) revealed a persistent but variegated profibrotic signature. In particular, we revealed a slight decrease in the expression of ADAMs (i.e., ADAM17, ADAMTS7, ADAMTS16), collagen genes (e.g., COL1A1, COL4A6) and the TGF-β receptor 1 gene (TGFBR1) in µDys-treated DMD organoids, when compared to the untreated condition, although still significantly different from isogenic control (Fig. 6E, right panel). Additionally, other fibrotic genes, like FN1 and COL1A1, integrins or cytokines (e.g.IL1A), showed no expression changes compared to untreated control (Fig. 6E, right panel). We further assessed the expression of key fibrotic genes identified by RNA-seq, including TGF-β, COL1A1, FN1, and ITGA10, in the other DMD MYOrganoids infected with uDys, alongside control DMD MYOrganoids infected with the empty vector to ensure the specific effect exerted by the transgene. Our analysis revealed that uDys gene transfer did not reduce the expression of TGF-β, COL1A1, FN1, and ITGA10, indicating the persistent expression of profibrotic genes in DMD-treated MYOrganoids (Fig. 6F). To understand the mechanisms behind the unresolved fibrotic process, we evaluated the reduction in TGF-β pathway activation within both cell subpopulations in DMD MYOrganoids following µDys treatment. To this aim, we generated organoids combining iPSC-derived myotubes (MYHC + ) and GFP-labeled fibroblasts. Histological staining revealed a significant decrease in pSMAD3-positive nuclei within the skeletal muscle subpopulation of µDys-treated DMD organoids, whereas no difference was observed in fibroblasts after µDys treatment as compared to untreated conditions (Fig. 6G). However, in both DMD myotube and fibroblast populations, µDys treatment did not lead to a fibrotic rescue, as shown by the significant differences in the pSMAD% versus CTR MYOrganoid populations. This result suggests a partial cell-autonomous correction of this signaling within the muscle population receiving µDys, but no paracrine effect on fibroblast activity. The persistence of activated TGF-β signaling in the myotube and fibroblast subpopulation after µDys gene transfer (Figs. 3I, J and 6G) can therefore explain the uncorrected profibrotic signature and poses the basis for future investigation aimed at improving or driving beneficial cell–cell communication.
Discussion
Here, we present a novel human-organoid model for DMD that simulates key traits of advanced stages, thanks to the strategic integration of fibroblasts and allows the interrogation of gene replacement effect on muscle function and fibrosis. Specifically, we report on the generation of 3D-engineered muscles called MYOrganoids, composed of a homogeneous population of iPSC-derived skeletal muscle cells30,31 and fibroblasts, major players in muscle tissue organization and microenvironment regulation34,35,67. We proved that fibroblast inclusion in the MYOrganoids results in improved structural maturation and functional performance under muscle force evaluation, while also eliciting a fibrotic signature in a DMD context.
Importantly, fibroblast incorporation was pivotal in exacerbating DMD phenotype and revealing fatiguability and muscle force loss, which is a top hallmark for DMD muscle function, thereby enabling its unprecedented use as therapeutic readouts in vitro. This is important considering that the capacity to evaluate muscle function in a high-throughput manner remains limited, further hindering progress in testing or developing effective treatments. As such, while several remarkable studies reported on muscle force in 3D models28,29,53,68,69, evaluating muscle function still presents challenges due to the complexity of identifying disease-specific force parameters and the intra-variability of in vitro models.
Furthermore, our study sheds new light on the mechanism underlying the contribution of fibroblasts to the modeling of DMD phenotype. The observation that both cell-contact and paracrine signaling are required to induce pathogenic modeling in DMD MYOrganoids indicates a direct crosstalk between fibroblasts and myofibers that can be further investigated to dissect mechanisms of action that sustain fibrosis independently of immune drivers. It also supports the potential of leveraging fibroblasts in tissue engineering as a source of ECM and profibrotic cues under pathological conditions.
Interestingly, DMD fibroblasts, when co-cultured with healthy myotubes, significantly increased the myofatiguability, while healthy fibroblasts mitigated the muscle performance of DMD myotubes. These findings indicate that fibrotic remodeling and excessive ECM protein deposition by DMD fibroblasts compromise the contractile performance of skeletal muscle cells, primarily via cell–cell contact-dependent mechanisms, unlike fibroblasts that appear to be refractory to dystrophic cues.
Overall, the exacerbated severity of DMD MYOrganoids enabled the evaluation of the therapeutic potential of µDys, currently employed in clinical trials. Consistent with the partial efficacy in the clinics13,14, µDys delivery in DMD MYOrganoids improved muscle force but did not rescue DMD phenotype, as shown by incomplete restoration of the components of the dystroglycan complex and limited reduction in fibrotic genes transcriptome. While direct evidence of DGC restoration in human patients post-gene transfer remains limited, aside from observations on beta-sarcoglycan restoration, which is less disrupted by dystrophin loss compared to alpha- and beta-dystroglycan70, our data confirms that μDys transfer in DMD MYOrganoids promotes functional improvement and partial effect on DGC rescue. Additionally, we observed a partial reduction in the TGF-β pathway activation exclusively on muscle cells, while no effect was observed in fibroblasts. This can explain the persistence of ECM protein production and secretion in µDys-treated DMD MYOrganoids, while we observed an effect in the restoration of muscle strength. These findings align with the evidence that mixed organoids, where DMD fibroblasts are co-cultured with either healthy or DMD myotubes, maintain increased secretion of ECM protein. This indicates that fibroblasts contribute to the profibrotic microenvironment autonomously and independently of dystrophin-deficient muscle signaling, suggesting the need to combine antifibrotic treatment that can synergistically improve outcomes of dystrophin restoration in DMD.
These findings also correlate with previous results showing that μDys exhibits limited effectiveness in reducing fibrosis in advanced disease stages71 and in more severe models and muscles affected by the disease, such as the diaphragm in the Dba2 mdx model72. We can speculate that, in advanced DMD, there may be a “point of no return” where the fibrotic program in fibroblasts becomes self-sustaining, due to persistent activation of pro-fibrotic signaling loops or altered cell–cell and cell–matrix interactions. Indeed, the stiffened ECM perpetuates fibrosis through mechanotransduction, activating fibroblasts to deposit ECM proteins and increasing rigidity and affecting skeletal muscle functions. This feedback loop hinders µDys’s ability to reverse fibrosis. This mechanistic insight helps explain the partial efficacy observed in human clinical trials, highlighting fibrosis as a major unresolved challenge in DMD treatment.
A limitation of in vitro human systems is undoubtedly the lack of the physiological complexity of an in vivo muscle. However, controlled cellular and environmental composition in MYOrganoids can still reproduce key hallmarks of human DMD phenotype and be suitable as a fast-screening platform for molecular studies and proof of concept data in a human genomic context, a task not feasible in animals. While additional cell types could be included, our direct myogenic conversion model and inclusion of a fibrotic source reproduce the advanced stage scenario of muscle decline, fibrosis and lack of regenerative potential. Although a certain degree of variability is expected in the therapeutic response across different patients due to the intrinsic heterogeneity of DMD patients73,74, MYOrganoids constitute a tailored system for testing dystrophin restoration in a severe context, with potential clinical relevance for personalized therapies.
Overall, pro-fibrotic DMD MYOrganoids provide a valuable tool to bridge the translational gap, either for the investigation of mechanisms driving the dystrophic process and as a human in vitro counterpart to animal in vivo preclinical studies, with the potential to accelerate the identification of new treatments.
Methods
Cell culture
Human iPSCs used in the study were as follows: CTR1 and CTR2 from healthy individuals, DMDdEx45, DMDdEx8-43 from DMD patients. The isogenic iPSC DMDdEx8-9 and corrected control (CRISPR-mediated skipping of mutated exons dEx6-9, isogenic control) were obtained from Olson lab53. The CTR1 and DMDdEx45 hiPSC lines were generated from skin fibroblasts from Coriell and were obtained from Marseille Stem Cells platform in Marseille Medical Genetics laboratory75. iPSCs were maintained using mTeSRplus medium and passaged using ReleSR (STEMCELL Technologies) on Matrigel-coated wells (Corning). iPSC engineering and muscle differentiation were performed by adapting a transgene-based method previously described31,32. All iPSC lines are engineered for transgene-based myotubes differentiation. Briefly, cells were nucleofected (4D Nucleofector, Lonza) with two enhanced versions of piggyBAC (ePB) containing respectively MyoD and BAF60c genes under tetracycline responsive promoter31,32. Additionally, iPSCs were regularly selected for the presence of the two ePBs with puromycin (10 µg/ml) and blasticidin (20 µg/ml) at the same time (Sigma-Aldrich). Human immortalized fibroblasts from control and DMD patients were generated and obtained from Myobank-AFM of Myology Institute from muscle biopsies. Control and DMD human immortalized fibroblasts, used in co-culture with hiPSC in 3D MYOrganoids, were maintained in culture in DMEM High Glucose Lglut supplemented with 20% fetal bovine serum (FBS) and passaged using TrypleE (Gibco). Human primary fibroblasts from control DMD patients were obtained from DNA bank of Genethon. Fibroblasts were maintained in DMEM high glucose (Gibco) supplemented with 20 ng/ml recombinant human fibroblast growth factor (h-FGF) (Peprotech) and 20% fetal bovine serum (Gibco). Primary fibroblasts were passed using TrypleE (Gibco). All cell lines are listed in table S5.
Generation of MYOrganoids
MYOrganoids from iPSC were generated adapting the protocol described for engineered heart tissue76. First, iPSC were induced with doxycycline 200 ng/µl for 24 hours to induce the expression of MyoD and Baf60c. The day after, 1.25 × 106 iPSC-committed were resuspended in 77 µl of growth media (SKM02, AMSbiokit) supplemented with hES cell Recovery (Stemgent) and molded in a hydrogel composed by 40 µl of Bovine Collagen solution 6 mg/ml (Sigma-Aldrich), 17.8 µl of Matrigel Growth Factor reduced (GFR, Corning) 10% v/v 3), 40 µl 2× DMEM (Gibco) 4) and 5.2 µl of NaOH 0.05 N). For the generation of MYOrganoids, including fibroblasts, 1.25 × 105 (ratio 1:10) fibroblasts were included in the iPSC-committed mix during hydrogel preparation. 180 µl of hydrogel was cast into a 48-well plate TM5 MyrPlate (Myriamed), containing in each well a pair of flexible poles (static stretchers) that support the growth of the engineered tissue in a ring shape. After 1 hour of polymerization at 37 °C, growth media (SKM02, AMSbiokit) were added for 24 hours. On day 2 of the 3D, growth media (SKM02, AMSbiokit) were replaced by differentiation media (SKM03plus, AMSbiokit) and changed every day until day 14. For Condition Media (CM) experiments, single-cell population organoid-like structures were made using the same protocol described above. CM was collected from CTR or DMD fibroblasts-only 3D structure and diluted 1:10 in fresh differentiation media (SKM03plus, AMSbiokit). The diluted media was then used to culture CTR or DMD myotubes-only organoids. CM was collected and used to treat 3D cultures from day 7 until day 13.
Muscle force analysis
Functional analyses were carried out on day 14 after 3D casting. Contraction experiments were performed using the MyoDynamics Muscle Strip System 840 MD (Danish Myo Technology A/S) and CS4 stimulator (Danish Myo Technology A/S). All functional analyses were performed at 37 °C, 5% CO2 95% O2, in Ringer solution at pH 7.1. Optimal muscle length (Lo) was determined by gradually stretching the muscle until 1.0 mN of passive tension was registered. Functional tests were performed under isometric and eccentric conditions. MYOrganoids were electrically stimulated with 250 pulses of 30 V, 4 ms width at the 125 Hz frequency for 2 seconds trains, for both isometric and eccentric contractions. For eccentric analysis, MYOrganoids were subjected to 1 mm stretch at the 6.5 mm/s speed during the muscular contraction, with 1 minute test between each repetition. Data collection and analysis were done by the PowerLab device and LabChart software (ADInstruments, New Zealand), respectively. Before the start of the protocol, 10 minutes recovery after the first isometric is given to prepare the muscle55. The contractility exercise was composed of 1 isometric contraction (Iso1), 10 eccentric contractions with 1 minute of rest between each repetition, and 1 last isometric contraction (Iso2). Fatigue index is represented as a percentage muscle force drop between Iso2-Iso1. Where indicated, force is indirectly normalized for the CSA (Cross Section Area) calculated as muscle force (mN) × Lo (mm) × density (mg/mm3)/weight (mg) and expressed as mN/mm2. Engineered muscle density is experimentally determined as 1.16984 mg/mm3.
Generation and isolation of fibroblasts expressing GFP
CTR (AB1191C16PV) and DMD (AB1024DMD11Q) immortalized fibroblasts were engineered in order to stably express GFP. Cells were infected with a lentivirus containing the GFP-cDNA under the control of a CMV promoter (LV-GFP). Briefly, cells were seeded at the density of 50,000 cells/mm2 and after 24 h have been infected with 2 × 107 IG/mL LV-GFP in the presence of the poloxamer Lentiboost (0.5 mg/mL) (Revvity) to enhance transduction. After 6 h, infection media was changed and replaced with fresh fibroblasts media. After 5 days, cells were detached with TrypleE Express 1× (Gibco) and then FACS-sorted with ASTRIOS-EQ (Beckman Coulter Life Sciences). Around 500,000 cells were sorted per condition, and then plated again in pre-warmed fibroblast media. Fibroblasts expressing GFP were then used to generate an organoid for pSMAD staining.
Immunofluorescence
MYOrganoids that did not undergo functional analysis have been analyzed for immunohistochemistry. Briefly, the artificial tissues were fixed in 4% methanol-free paraformaldehyde (PFA) overnight on day 14. For staining on transversal or longitudinal sections, fixed MYOrganoids were dehydrated with a gradient of sucrose (7.5%–30%) over day and embedded in OCT matrix in plastic molds. After 24 hours, embedded MYOrganoids were processed with the cryostat (LEICA) with 12 µm thick sections. Slices were then dried and fixed again with 4% methanol-free PFA at RT for 10 minutes (Invitrogen). Fixed sections were then blocked with serum cocktail (5% Goat serum and 5% Fetal bovine serum), before being stained overnight at +4 °C with primary antibody. After that, slices were washed three times in PBS and hybridized with AlexaFluor secondary antibody according to the host species of the first antibody. Stained slides were then covered with Fluoromont + Dapi (SouthernBiotech) and glass slide 1.5H. For imaging, sections were scanned with AxioScan microscope and confocal Leica SP8. For 2D staining, cells were grown on µ-Dish 35 mm (Ibidi) and then fixed in 4% methanol-free PFA for 7 minutes. For membrane and cytoplasmic staining, cells were permeabilized with 0.15% Triton X-100 for 10 minutes and then washed with PBS for 5 minutes. For nuclear staining, cells were permeabilized with 0.25% Triton X-100 for 15 minutes. Permeabilized cells were then blocked with serum cocktail (5% Goat serum and 5% Fetal bovine serum), before being stained overnight at +4 °C with primary antibody. Cells are then washed three times with PBS and then hybridized with AlexaFluor secondary antibody according to the host species of the first antibody. After three PBS washes, nuclei were stained with Hoechst 33342 (Invitrogen) at the final dilution of 2 µg/ml. For imaging, sections are scanned confocal Leica SP8. All antibodies used for staining are listed in table S6.
Electron microscopy
Electron microscopy analysis was prospectively performed on MYOrganoids specimens that were fixed with glutaraldehyde (2.5%, pH 7.4), post-fixed with osmium tetroxide (2%), dehydrated in a graded series of ethanol ranging from 30% to absolute solution and embedded in resin (EMBed-812, Electron Microscopy Sciences, USA). 80 nm thick sections from at least four blocks from CTR iPSC-derived MYOrganoids in the presence or absence of CTR fibroblasts were stained with uranyl acetate and lead citrate. The grids were observed using a “JEOL 1400 Flash” electron microscope (120 kV) and were photo documented using a Xarosa camera (Soft Imaging System, France). Images covering whole longitudinal sections were assessed, and representative images were used.
Images analysis
Myotubes alignment: FIJI and CellPose were used for image analysis. Myotube alignment was determined by angle measurement and by myotube circularity from cross-section cuts of MYOrganoids. Angles between two myotubes were measured with the “angle tool” function in FIJI, by drawing two lines perpendicular to two adjacent myotubes' membranes stained with WGA. Myotubes circularity was determined by custom FIJI script. Briefly, myotubes cross-section area was first segmented by pre-trained Cellpose2 cyto2 model77 and then converted into Regions of Interest (ROIs) by Labels_To_Rois.py plugin (Waisman et al. 2021) for subsequent quantification on FIJI (Ferret diameters X and Y). After that, the ratio between Feret’s diameter on axis X and axis Y was assessed. At least 4 frames for 3 biological replicates have been analyzed. Striation frequency and fusion index: Striation frequency was evaluated by counting striated myotubes, delineated by either sarcomeric alpha-actinin (SAA) and membrane marker WGA staining, and normalized for the total number of nuclei contained in myotubes, using FIJI software. Fusion index has been calculated by counting myotubes myosin heavy chain (MYHC) positive myotubes containing 1, 2, 3 or more than 3 nuclei as a percentage of the total number of nuclei. At least 4 frames for 3 biological replicates have been analyzed. Z-disk length analysis: Z-disk length has been assessed by drawing straight lines in FIJI and by measuring the length in at least 10 mature myotubes per condition. Phospho-SMAD3 quantification: Phospho-SMAD3 (pSMAD3) has been measured as a percentage of positive nuclei of myosin heavy chain positive cells (MyHC + , i.e., myotubes) or GFP-positive cells (GFP+ i.e., fibroblasts). For each condition, 4 frames for 3 biological replicates have been analyzed. For this purpose, a custom FIJI macro has been generated. Briefly, ROIs have been defined based on DAPI staining (threshold: 70/255) and overlapped with pSMAD signal (threshold: 40/255), to calculate the number of pSMAD+ nuclei. After that, the same ROIs have been applied to GFP (threshold 40/255) and MYHC signals (threshold 40/255), to calculate fibroblast nuclei (GFP+) and myotube nuclei (MYHC+). Each ROI was considered positive with a mean fluorescence > 5 for pSMAD, and >0 for GFP and MYHC. Dystrophin, α-Dystroglycan and β-Dystroglycan quantification: MYOtissue cross sections were stained for sarcomeric alpha-actinin (SAA) for myotube cytosol labeling. The Cellpose2 cyto2 model 10 was fine-tuned on manually myofiber-labeled images based on SAA staining (hyperparameters: n_epochs = 200, learning_rate = 0.05, weight_decay = 0.0001). The labeled dataset used in fine-tuning was prepared in such a way that the model can simultaneously segment myotubes and ignore low-quality staining areas. Fine-tuned models were then used to extract myotube masks. Reconstruction of myotube masks was done using the cellpose package (Stringer et al. 2021). Reconstructed masks were then converted into Regions of Interest (ROIs) for subsequent quantification (each ROI corresponds to an individual myofiber) using the Labels_To_Rois.py FIJI plugin78. The generated ROIs were used for subsequent quantification using FIJI macro. Positive ROIs for dystrophin / alpha-dystroglycan/beta-dystroglycan signal were counted and represented as a percentage of the total ROIs of one image.
Gene expression analysis
For gene expression analysis, bulk RNA was isolated from MYOrganoids by RNeasy micro kit (QIAGEN) according to manufacturer’s instructions, controlled and quantified by Nanodrop. Around 0.5–1 µg of RNA was retro-transcribed to cDNA thanks to the RevertAid H Minus First Strand cDNA Synthesis Kit (Invitrogen). Droplet digital PCR was performed to assess the expression of myogenic factors (MYOGENIN, MYH2, MYH7, MCK), of fibrotic markers (COL1A1, FN1, TGF-β1) and µDystrophin, thanks to the QX200™ ddPCR™ EvaGreen Supermix (Biorad). Gene expression results in copy/µl (cp/µl) are represented as fold change to the expression of GAPDH gene. Primers used are listed in table S7.
Evans blue dye (EBD) assay
To measure the membrane permeability after eccentric contraction, we performed EBD staining. Briefly, MYOrganoids were subjected to 1 eccentric contraction and then placed in the incubator with a solution composed of 0.25 mg/ml of EBD diluted in differentiation media for 1 hour. As a control, organoids were treated with 6 µg/ml Notexin for 1 hour, washed 3 times in PBS and then fixed overnight with methanol-free PFA (4%) at +4 C. Staining was performed as previously described.
ELISA assay
Media was collected before muscle functional analysis after being in culture for 48 hours. Secreted human Fibronectin 1 (FN1), TGF-β and Collagen IV were assessed by ELISA kit (table S8) from MYOrganoids supernatant.
AAV production and MYOrganoids infection
Recombinant AAVs were produced as previously described (Bourg et al. 2022) using AAV9 serotype. Purification was performed using affinity chromatography, and titration was done by ddPCR using transgene-specific primers. For optimization of infection, an AAV9-CMV-GFP construct was used. The micro-dystrophin transgene used in the study, under the control of spc512 promoter, was an optimized version of the construct used for GENETHON’s preclinical investigation and clinical trial14, with deletion from spectrin-like repeats 4–23 and full C-terminal truncation, here referred to as µDys. Infection in MYOrganoids was performed by delivering the AAV9 particles diluted into the differentiation media at day 7, at two different doses: 1E + 9 vg/MYOtissue (low dose) and 5E + 10 vg/MYOtissue (high dose). Media was replaced after 24 hours from the infection and changed daily until day 14.
Viral copy number analysis
Viral DNA was extracted from mature MYOrganoids by NucleoMag Pathogen kit (Macherey Nagel) using Kingfisher instrument (Thermofisher). DNA yield and purity were assessed by Nanodrop; viral copy number (VCN) was identified by droplet digital PCR using supermiral copy numix for probe (Biorad). Results are shown as copy number variation using P0 as the reference DNA. Primers used are listed in table S7.
Capillary western blot analysis
MYOrganoids proteins were extracted in RIPA buffer supplemented with Protease Inhibitor Cocktail EDTA-free (Roche) and Benzonase by homogenization. Total proteins were then quantified by BCA method, thanks to the Pierce 660 protein assay kit (Invitrogen) according to manufacturer’s instructions. Protein detection has been performed by capillary western blot, thanks to the JESS protein simple 66–440 kDa module (Bio-techne), according to the manufacturer’s directions. For dystrophin (both full-length and µDystrophin) and pSMAD detection, 2.5 μg of total protein was loaded in each well. A chemiluminescent signal was detected and quantified automatically by SW Compass software and showed as a corrected area normalized on total protein. List of antibodies used for capillary western blot is available in Table S6. Unprocessed capillary western blot lanes and respective chemiluminescence peaks are deposited in Fig. S11.
RNA sequencing and transcriptomic analysis
The RNA quality of samples was verified using the Bioanalyzer 2100 (Agilent) and Qubit fluorometric quantification (ThermoFisher Scientific). The samples that had an RNA integrity number higher than 9 were used for RNA sequencing (Genewiz). The Stranded Total RNA Library Prep Kit (Illumina) was used to create sequencing libraries, which were sequenced following the Illumina protocol on the NovaSeq instrument (Illumina), resulting in approximately 20 million paired-end reads per library. The paired-end reads were filtered and subjected to quality control using fastp (Chen et al. 2018). They were then mapped to the GRCh38/hg38 genome using HISAT279 count tables were generated using htseq-count80. Differentially expressed genes (DEGs) were identified using the DESeq2 R package with p value adjusted by Benjamin–Hochberg procedure less than 0.05. Pathway analysis was carried out in R-Studio (version 4.0.3) using functional class scoring with Gene Set Enrichment Analysis81,82.
Statistical analysis
For comparisons between two independent groups, an unpaired, one-tailed Student’s t test was applied when a directional hypothesis was specified. To compare multiple groups, we used one-way analysis of variance with Tukey’s, Šidák's, or Dunnett’s correction for multiple comparison tests, with the assumption of Gaussian distribution of residuals. Results were considered significantly different at p < 0.05. Statistical analysis and graphs were realized by GraphPad Prism v10.4.1 software and R version 2025.05.0. The figures display the mean ± standard error of the mean. N indicates the number of biological replicates, as described in the figure legend.
Data availability
The data supporting the findings of this study are available within the article and its supplemental information. The RNA sequencing datasets generated in this study can be found in the NCBI Bioproject database (https://www.ncbi.nlm.nih.gov/bioproject) using the access number PRJNA1208956. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sonia Albini (salbini@genethon.fr).
References
Mendell, J. R. et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 71, 304–313 (2012).
Hoffman, E. P., Brown, R. H. & Kunkel, L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987).
Ervasti, J. M. & Campbell, K. P. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 122, 809–823 (1993).
Petrof, B. J., Shrager, J. B., Stedman, H. H., Kelly, A. M. & Sweeney, H. L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA 90, 3710–3714 (1993).
Hoffman, E. P. The discovery of dystrophin, the protein product of the Duchenne muscular dystrophy gene. FEBS J. 287, 3879–3887 (2020).
Davies, K. E. & Nowak, K. J. Molecular mechanisms of muscular dystrophies: old and new players. Nat. Rev. Mol. Cell Biol. 7, 762–773 (2006).
Ennen, J. P., Verma, M. & Asakura, A. Vascular-targeted therapies for Duchenne muscular dystrophy. Skelet Muscle 3, 9 (2013).
Porter, J. D. et al. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum. Mol. Genet. 11, 263–272 (2002).
Amor, F. et al. Cholesterol metabolism is a potential therapeutic target in Duchenne muscular dystrophy. J. Cachexia Sarcopenia Muscle 12, 677–693 (2021).
Mendell, J. R. et al. duan. Mol. Ther. 29, 464–488 (2021).
Aguti, S., Malerba, A. & Zhou, H. The progress of AAV-mediated gene therapy in neuromuscular disorders. Expert Opin. Biol. Ther. 18, 681–693 (2018).
Duan, D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol. Ther. 26, 2337–2356 (2018).
Le Guiner, C. et al. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun. 8, 16105 (2017).
Elangkovan, N. & Dickson, G. Gene therapy for Duchenne muscular dystrophy. J. Neuromuscul. Dis. 8, S303–S316 (2021).
Harper, S. Q. et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 8, 253–261 (2002).
Wang, B., Li, J. & Xiao, X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc. Natl. Acad. Sci. USA 97, 13714–13719 (2000).
Fabb, S. A., Wells, D. J., Serpente, P. & Dickson, G. Adeno-associated virus vector gene transfer and sarcolemmal expression of a 144 kDa micro-dystrophin effectively restores the dystrophin-associated protein complex and inhibits myofibre degeneration in nude/mdx mice. Hum. Mol. Genet. 11, 733–741 (2002).
Duan, D. Duchenne muscular dystrophy gene therapy in 2023: status, perspective, and beyond. Hum. Gene Ther. 34, 345–349 (2023).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Tornabene, P. et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 11, eaav4523 (2019).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Hofer, M. & Lutolf, M. P. Engineering organoids. Nat. Rev. Mater. 6, 402–420 (2021).
Garita-Hernandez, M. et al. AAV-mediated gene delivery to 3d retinal organoids derived from human induced pluripotent stem cells. Int. J. Mol. Sci. 21, 994 (2020).
Kruczek, K. et al. Gene therapy of dominant CRX-leber congenital amaurosis using patient stem cell-derived retinal organoids. Stem Cell Rep. 16, 252–263 (2021).
Khabou, H. et al. Noninvasive gene delivery to foveal cones for vision restoration. JCI Insight. 3, e96029 (2018).
Shin, M. K. et al. Generation of skeletal muscle organoids from human pluripotent stem cells to model myogenesis and muscle regeneration. Int. J. Mol. Sci. 23, 5108 (2022).
Pinton, L. et al. 3D human induced pluripotent stem cell-derived bioengineered skeletal muscles for tissue, disease and therapy modeling. Nat. Protoc. 18, 1337–1376 (2023).
Rao, L., Qian, Y., Khodabukus, A., Ribar, T. & Bursac, N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 9, 126 (2018).
Shahriyari, M. et al. Engineered skeletal muscle recapitulates human muscle development, regeneration and dystrophy. J. Cachexia Sarcopenia Muscle 13, 3106–3121 (2022).
Caputo, L. et al. Acute conversion of patient-derived Duchenne muscular dystrophy iPSC into myotubes reveals constitutive and inducible over-activation of TGFβ-dependent pro-fibrotic signaling. Skelet. Muscle 10, 13 (2020).
Albini, S. et al. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 3, 661–670 (2013).
Albini, S. & Puri, P. L. Generation of myospheres from hESCs by epigenetic reprogramming. J. Vis. Exp. e51243 (2014).
Iberite, F., Gruppioni, E. & Ricotti, L. Skeletal muscle differentiation of human iPSCs meets bioengineering strategies: perspectives and challenges. NPJ Regen. Med. 7, 23 (2022).
Mathew, S. J. et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138, 371–384 (2011).
Bersini, S. et al. Engineering an environment for the study of fibrosis: A 3D human muscle model with endothelium specificity and endomysium. Cell Rep. 25, 3858–3868.e4 (2018).
Tiburcy, M., Meyer, T., Soong, P. L. & Zimmermann, W. H. Collagen-based engineered heart muscle. Methods Mol. Biol. 1181, 167–176 (2014).
Rao, N. et al. Fibroblasts influence muscle progenitor differentiation and alignment in contact independent and dependent manners in organized co-culture devices. Biomed. Microdevices 15, 161 (2013).
Saleh, K. K. et al. Single cell sequencing maps skeletal muscle cellular diversity as disease severity increases in dystrophic mouse models. iScience 25, 105415 (2022).
Giordani, L. et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 (2019).
Petrany, M. J. et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 11, 6374 (2020).
Karlsen, A. et al. Distinct myofibre domains of the human myotendinous junction revealed by single-nucleus RNA sequencing. J. Cell Sci. 136, jcs260913 (2023).
Thorsteinsdottir, S., Deries, M., Cachaço, A. S. & Bajanca, F. The extracellular matrix dimension of skeletal muscle development. Dev. Biol. 354, 191–207 (2011).
Costantini, M. et al. Biofabricating murine and human myo-substitutes for rapid volumetric muscle loss restoration. EMBO Mol. Med. 13, e12778 (2021).
Al Tanoury, Z. et al. Prednisolone rescues Duchenne muscular dystrophy phenotypes in human pluripotent stem cell-derived skeletal muscle in vitro. Proc. Natl. Acad. Sci. USA 118, (2021).
Incitti, T. et al. Pluripotent stem cell-derived skeletal muscle fibers preferentially express myosin heavy-chain isoforms associated with slow and oxidative muscles. Skelet. Muscle 10, 17 (2020).
Pette, D. & Staron, R. S. Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509 (2000).
Minato, K. et al. Measurement of lateral transmission of force in the extensor digitorum longus muscle of young and old mice. Int. J. Mol. Sci. 22, 12356 (2021).
Erdogan, B. R. et al. Normalization of organ bath contraction data for tissue specimen size: does one approach fit all? Naunyn Schmiedebergs Arch. Pharm. 393, 243–251 (2020).
Santos, G. L. et al. Fibroblast derived human engineered connective tissue for screening applications. J. Vis. Exp. 2021 (2021).
Wynn, T. A. Cellular and molecular mechanisms of fibrosis J. Pathol. 214, 199–210 (2008).
Kharraz, Y., Guerra, J., Pessina, P., Serrano, A. L. & Muñoz-Cánoves, P. Understanding the process of fibrosis in Duchenne muscular dystrophy. Biomed. Res. Int. 2014, 965631 (2014).
Mann, C. J. et al. Aberrant repair and fibrosis development in skeletal muscle. Skelet. Muscle 1, 21 (2011).
Kyrychenko, V. et al. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI Insight 2, e95918 (2017).
Childers, M. K. et al. Eccentric contraction injury in dystrophic canine muscle. Arch. Phys. Med Rehabil. 83, 1572–1578 (2002).
García-Sillero, M. et al. Comparison of interventional strategies to improve recovery after eccentric exercise-induced muscle fatigue. Int. J. Environ. Res. Public Health 18, 1–11 (2021).
Hu, X. et al. A novel ex vivo protocol that mimics length and excitation changes of human muscles during walking induces force losses in EDL but not in soleus of mdx mice. PLoS One 20, e0320901 (2025).
Allen, D. G., Lamb, G. D. & Westerblad, H. Skeletal muscle fatigue: Cellular mechanisms. Physiol. Rev. 88, 287–332 (2008).
Peczkowski, K. K. et al. Muscle twitch kinetics are dependent on muscle group, disease state, and age in Duchenne muscular dystrophy mouse models. Front. Physiol. 11, 568909 (2020).
Klingberg, F. et al. The fibronectin ED-A domain enhances recruitment of latent TGF-β-binding protein-1 to the fibroblast matrix. J. Cell. Sci. 131, jcs201293 (2018)
Zanotti, S., Gibertini, S. & Mora, M. Altered production of extra-cellular matrix components by muscle-derived Duchenne muscular dystrophy fibroblasts before and after TGF-beta1 treatment. Cell Tissue Res. 339, 397–410 (2010).
Weber, K. T., Sun, Y., Bhattacharya, S. K., Ahokas, R. A. & Gerling, I. C. Myofibroblast-mediated mechanisms of pathological remodelling of the heart. Nat. Rev. Cardiol. 10, 15–26 (2013).
Giuliani, G., Rosina, M. & Reggio, A. Signaling pathways regulating the fate of fibro/adipogenic progenitors (FAPs) in skeletal muscle regeneration and disease. FEBS J. 289, 6484–6517 (2022).
Manini, A., Abati, E., Nuredini, A., Corti, S. & Comi, G.P. Adeno-associated virus (AAV)-mediated gene therapy for duchenne muscular dystrophy: the issue of transgene persistence. Front. Neurol. 12, 814174 (2021).
Pacak, C. A. et al. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 99, e3-9 (2006).
Muraine, L. et al. Transduction efficiency of adeno-associated virus serotypes after local injection in mouse and human skeletal muscle. Hum. Gene Ther. 31, 233–240 (2020).
Lapidos, K. A., Kakkar, R. & McNally, E. M. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ. Res. 94, 1023–1031 (2004).
Li, M., Dickinson, C. E., Finkelstein, E. B., Neville, C. M. & Sundback, C. A. The role of fibroblasts in self-assembled skeletal muscle. Tissue Eng. Part A 17, 2641–2650 (2011).
Madden, L., Juhas, M., Kraus, W. E., Truskey, G. A. & Bursac, N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife 4, e04885 (2015).
Khodabukus, A. et al. Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials https://doi.org/10.1016/j.biomaterials.2018.08.058 (2018).
Capitanio, D. et al. Comparative proteomic analyses of Duchenne muscular dystrophy and Becker muscular dystrophy muscles: changes contributing to preserve muscle function in Becker muscular dystrophy patients. J. Cachexia Sarcopenia Muscle 11, 547–563 (2020).
Heller, K. N., Mendell, J. T., Mendell, J. R. & Rodino-Klapac, L. R. MicroRNA-29 overexpression by adeno-associated virus suppresses fibrosis and restores muscle function in combination with micro-dystrophin. JCI Insight 2, e93309 (2017).
Hart, C. C. et al. Potential limitations of microdystrophin gene therapy for Duchenne muscular dystrophy. JCI Insight 9, e165869 (2024).
Desguerre, I. et al. Clinical heterogeneity of Duchenne muscular dystrophy (DMD): definition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS One 4, e4347 (2009).
Towse, T. F. et al. Post-contractile BOLD contrast in skeletal muscle at 7 T reveals inter-individual heterogeneity in the physiological responses to muscle contraction. NMR Biomed. 29, 1720–1728 (2016).
Badja, C. et al. Efficient and cost-effective generation of mature neurons from human induced pluripotent stem cells. Stem Cells Transl. Med. 3, 1467–1472 (2014).
Tiburcy, M., Meyer, T., Liaw, N. Y. & Zimmermann, W. H. Generation of engineered human myocardium in a multi-well format. STAR Protoc. 1, 100032 (2020).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Waisman, A., Norris, A. M., Elías Costa, M. & Kopinke, D. Automatic and unbiased segmentation and quantification of myofibers in skeletal muscle. Sci. Rep. 11, 11793 (2021).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Acknowledgements
We express our gratitude to Rene Hummel (Danish MyoTechnology), Guillaume Tanniou, and Nicolas Guerchet for their technical support in muscle force evaluation. We thank Guillaume Corre for the aid with RNA-seq analysis. We thank the service of bioproduction of Genethon for preparing the AAV particles used in this study. We also extend our appreciation to Jérémie Cosette and Peggy Sanatine for the assistance in imaging and cell-sorting, and to the histology team for their technical expertise. Finally, we would like to thank Frederique Magdinier for generously providing the DMD iPSC dEx45 and CTR1 iPSC. This study was financially supported by the Institut National de la Sante et de la Recherche Medicale (INSERM), by ANR 23-CE52-0003, and by the “Association Française contre les Myopathies” (AFM).
Author information
Authors and Affiliations
Contributions
S.A. and LP.a designed the experiments. L.Pa performed most of the experiments, including iPSC culture, organoid generation, imaging, and muscle force analysis. M.F. performed capillary western blot analysis and ELISA assays. L.Pi performed force assays and gene and protein expression analysis. G.Bi performed organoid generation and conditioned media experiments. M.F. and A.J. performed secretome analysis. M.M. optimized muscle force assays. A.V.H. analyzed RNA-seq data. R.E.K. and G.Br performed electron microscopy analysis. A.B. generated and provided the immortalized human fibroblasts. S.A. and L.Pa performed data analysis and figure preparation. S.A. conceived, supervised the project, and wrote the manuscript. All authors discussed the results. S.A., L.Pa., D.I., and I.R. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Palmieri, L., Bimbi, G., Ferrand, M. et al. Disease exacerbation in human DMD MYOrganoids enables gene therapy evaluation and unveils persistence of fibrotic activity. npj Regen Med 11, 2 (2026). https://doi.org/10.1038/s41536-025-00445-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41536-025-00445-8