Abstract
MicroRNAs (miRNAs) are critical regulators of neurodevelopment and are implicated in the pathogenesis of schizophrenia. Schizophrenia is increasingly recognized as a neurodevelopmental disorder, with most cases emerging during late adolescence and early adulthood, which is a critical period of brain maturation. However, the study of miRNAs during this phase has been limited by the challenges of postmortem brain analysis. Neuron-derived extracellular vesicles (NEVs) have recently been proposed for investigating brain-derived molecular profiles. In this study, NEVs were enriched from plasma using the L1CAM antibody in patients with recent-onset schizophrenia (ROS) within 5 years of onsets and chronic-phase schizophrenia (CS). The miRNA profiles of these NEVs in patients with ROS and CS were compared with those of age-and sex-matched healthy controls. Differential expression analysis revealed miRNA changes specific to the recent-onset phase as well as possible pathophysiological mechanisms transitioning from the recent-onset to the chronic phases. These findings provide novel insights into the role of miRNAs in neurodevelopmental abnormalities associated with schizophrenia onset. This study highlights the utility of NEVs as a tool for accessing brain-derived miRNA profiles and diagnostic biomarkers and underscores the importance of an onset period as a critical window for understanding the molecular underpinnings of schizophrenia.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs, composed of 20–24 nucleotides that regulate gene expression by binding to mRNAs with complementary sequences, leading to their degradation or inhibition of protein translation1. They play crucial roles in various biological processes, including neurogenesis and synaptic connectivity, making them essential for brain development from the fetal stage to adolescence2,3.
Several studies have reported an association between miRNAs and schizophrenia4,5,6,7. The signs and symptoms of schizophrenia typically emerge during late adolescence and early adulthood, with approximately 60–70% of patients experiencing their first occurrence between the ages of 14 and 25 years and additional cases, particularly in early adulthood, extending into the late twenties8. While many patients develop the disorder during late adolescence, a substantial subset, particularly women, experience onset in early adulthood or even after the age of 309. Late adolescence and early adulthood are critical periods for brain maturation10,11,12,13, and schizophrenia is increasingly understood as a neurodevelopmental disorder14,15,16. Therefore, understanding brain processes during this developmental time window and onset period is essential for elucidating the pathogenesis related to the onset of schizophrenia16,17. Despite this, few studies have focused on miRNA dynamics during this pivotal period in the context of schizophrenia18.
Recently, neuron-derived extracellular vesicles (NEVs) have attracted significant attention as a novel tool for investigating brain physiology and pathology through peripheral samples19,20,21,22. Extracellular vesicles (EVs) are small phospholipid bilayer-enclosed vesicles that are abundant in body fluids such as blood and urine23,24. They originate from various cell types and carry proteins, nucleic acids, and metabolites, reflecting the molecular environment of their tissues of origin25,26,27. Moreover, EVs possess tissue-specific membrane proteins, enabling the identification of their cellular sources28. NEVs, in particular, can be enriched from plasma by immunoprecipitation using neuron-specific markers such as L1CAM and MAP1B antibodies29,30. Notably, EVs contain miRNAs31,32, making them promising tools for analyzing neuronal miRNA profiles using peripheral blood samples22,33.
In this study, we investigated the role of miRNAs in schizophrenia by enriching NEVs from the plasma of patients with recent-onset schizophrenia (ROS) within 5 years of onsets and chronic-phase schizophrenia (CS). We analyzed the miRNA profiles of NEVs from these patients and compared them with those from age- and sex-matched healthy controls. We conducted a functional analysis of differentially expressed miRNAs to uncover insights into their roles in both the ROS and CS. Furthermore, we evaluated the diagnostic potential of miRNAs as biomarkers for ROS.
Results
Demographic data and clinical characteristics of participants
This study analyzed NEVs enriched from the peripheral blood of 14 patients with schizophrenia within 5 years of onset (ROS group), 15 age- and sex-matched controls (CROS group), 8 patients with schizophrenia for more than 15 years post-onset (CS group), and 8 age- and sex-matched controls (CCS group). All the participants underwent miRNA sequencing.
The demographic data and clinical characteristics are summarized in Table 1. No significant differences in age or sex were observed between the ROS and CROS groups or between the CS and CCS groups. In the ROS group, nine patients were outpatients and five were inpatients, whereas in the CS group, two were outpatients and six were inpatients. Positive and Negative Syndrome Scale (PANSS) scores were available for 11 patients in the ROS group and all eight patients in the CS group. The mean total PANSS score in the ROS group was 38.2 ± 8.8, corresponding to mild symptom severity and indicating that these patients were not acutely psychotic at the time of sampling. By contrast, the mean total PANSS score in the CS group was 106.5 ± 12.4, reflecting persistent and more severe psychopathology typical of chronic schizophrenia. Importantly, none of the patients in either group received clozapine at the time of blood collection. The ROS group represented patients with ROS in early adulthood (mean age 27.5 years), reflecting a typical age of onset rather than an early-onset subgroup. Additionally, for some participants, information on tobacco and alcohol use, as well as medication details, was collected. Patients in both the ROS and CS groups were recruited from the Tokyo Metropolitan Matsuzawa Hospital, and all were receiving pharmacological therapy.
NEV enrichment and validation
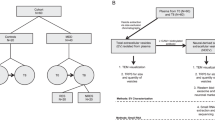
Plasma-derived EVs were first isolated using the ExoQuick reagent. These whole EV preparations were characterized in accordance with the Minimal Information for Studies of Extracellular Vesicles (MISEV) 2023 guidelines34, including nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM), to confirm the presence and morphology of EVs. NTA (Nanosight) revealed a size distribution ranging from 50 to 400 nm, with a peak size of 162 ± 9.9 nm, consistent with EVs (Fig. 1A). TEM confirmed the presence of EVs in plasma-derived samples (Fig. 1B).
A Size distributions of plasma-derived extracellular vesicles (EVs) from participants determined via nanoparticle tracking analysis. Measurements were performed five times. The vertical axis represents the number of particles per milliliter, and the horizontal axis indicates particle size. The black line represents the mean value, and the red error bars indicate 1 standard error of the mean. B Negative-staining transmission electron microscopy images of plasma-derived EVs from participants. The scale bar indicates 100 nm. C Characterization of NEV-enriched samples by marker protein antibody array. This array included two antibodies for exosome markers (CD81 and TSG101), three antibodies for neuronal markers (L1CAM, ENO2, MAPT), and one antibody for cellular contamination (CANX).
Following this validation, neuron-enriched vesicle (NEV) fractions were collected from the characterized EV preparations by immunocapture using an L1CAM antibody. Antibody array analysis revealed the presence of exosome marker proteins (CD81 and TSG101) and neuronal markers (L1CAM, ENO2, and MAPT), whereas the absence of an endoplasmic reticulum marker (CANX) confirmed minimal contamination from intracellular vesicles (Fig. 1C). These findings confirmed the successful NEV enrichment for downstream analysis.
Although previous studies have demonstrated that L1CAM+ fractions can be directly characterized by NTA and TEM35,36, the yield from our plasma samples was not sufficient for reliable measurements, and the elution process might have altered vesicle morphology. Therefore, in this study, we used these assays to validate the total EV population, from which L1CAM+ fractions were subsequently enriched for downstream molecular analyses.
Estimation of tissue origin for miRNAs in NEVs
Small RNA-seq identified 2,884 miRNAs across 45 samples, and 128 miRNAs detected in more than 75% of samples were retained for further analysis. These miRNAs included brain-specific miRNAs, such as miR-13437, miR-12737, and miR-12838. Tissue-specific enrichment analysis of miRNA target genes showed significant enrichment of genes expressed in brain regions, including the prefrontal cortex, midbrain, and cingulate gyrus, compared with other tissues (Table S1). These findings confirmed the neuronal origin of the miRNAs in the NEV samples.
miRNA differential expression analysis
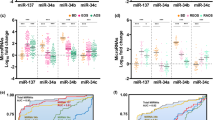
Seventeen miRNAs were differentially expressed between the ROS and CROS groups, meeting an FDR threshold <5% and a fold change >2 or <0.5 (Fig. 2A). Among these, 10 miRNAs (hsa-miR-122-5p, hsa-miR-511-5p, hsa-miR-483-5p, hsa-miR-192-5p, hsa-miR-99a-5p, hsa-miR-342-5p, hsa-miR-193a-5p, hsa-miR-200a-3p, hsa-miR-342-3p, and hsa-miR-375-3p) were upregulated, whereas 7 (hsa-miR-100-5p, hsa-miR-423-3p, hsa-miR-361-5p, hsa-miR-143-3p, miR-3184-5p, hsa-miR-584-5p, and hsa-miR-184) were downregulated (Table S2).
A Significant differential expression was observed in NEVs of patients with recent-onset schizophrenia (n = 14) compared with age- and sex-matched healthy controls (n = 15). B Significant differential expression was observed in NEVs of patients with chronic-phase schizophrenia (n = 8) compared with age- and sex-matched healthy controls (n = 8). The fold change of each miRNA is plotted against its corrected P value; blue circles represent miRNAs with FDR < 0.05, and log2FC < −1.0. Red lines represent the threshold of significant difference with FDR < 0.05, and |log2FC | > 1.0.
In the CS vs. CCS group comparison, only two miRNAs (hsa-miR-184 and hsa-let-7c-5p) were significantly downregulated (Fig. 2B, Table S3). Notably, hsa-miR-184 was consistently downregulated in both the ROS and CS groups compared with their respective controls.
Further analysis revealed associations between miRNA expression and clinical variables (Table S4). hsa-miR-122-5p was significantly associated with the dosage of benzodiazepine receptor agonist hypnotics, and hsa-miR-193a-5p was associated with antidepressant dosage.
Performance of miRNAs as biomarkers for recent-onset schizophrenia
We evaluated the potential of miRNAs as biomarkers for ROS, which in our sample corresponded to early adulthood rather than adolescence. Of the 17 differentially expressed miRNAs, two were excluded due to their association with medication dosage. We then performed univariate logistic regression of the remaining 15 miRNAs, with p values adjusted using the Benjamini–Hochberg false discovery rate (FDR) procedure, followed by the assessment of area under the curve (AUC). We found hsa-miR-143-3p showing the highest AUC score (0.933, 95% confidence interval (CI) = 0.803–1.00) and odds ratio (0.222, 95% CI = 0.0645–0.475) (Table 2). The sensitivity, specificity, and accuracy of hsa-miR-143-3p were 100 ± 0.00%, 93.3 ± 3.33%, and 96.6 ± 1.72%, respectively, where the thresholds were selected as 0.787 ± 0.253 based on the Youden index (Table S5).
Functional analysis of miRNA
NEVs likely influence gene expression in originating neurons and target cells via intercellular communication. To investigate the functional roles of miRNAs, target genes expressed in glutamatergic neurons, GABAergic neurons, and astrocytes were identified39.
Gene Ontology (GO) analysis revealed that the target genes of miRNAs differentially expressed in the ROS group were associated with neuron projection development and endosomal transport (Fig. 3A and Tables S5–S7). Conversely, miRNAs differentially expressed in the CS group were linked to the ER-nucleus signaling pathway and the regulation of postsynaptic membrane neurotransmitter receptor levels (Fig. 3B and Tables S8–S10). These findings suggest distinct functional effects of miRNAs in the onset and chronic phases of schizophrenia.
A Gene Ontology (GO) analysis of differentially expressed miRNAs’ target genes in the recent-onset group. The miRNAs were differentially expressed between participants with recent-onset schizophrenia and age- and sex-matched controls. B GO analysis of differentially expressed miRNAs’ target genes in the chronic-phase group. The miRNAs were differentially expressed between participants with chronic schizophrenia and age- and sex-matched controls. These analyses were conducted on target genes expressed in each cell type (glutamatergic neurons, GABAergic neurons, and astrocytes). The top 10 GO biological process terms ranked by enrichment ratio are presented. Blue bars represent GO terms with an FDR < 0.05. Light blue bars represent GO terms with FDR > 0.05.
Discussion
This study demonstrates the utility of plasma NEVs for investigating neuronal miRNA expression profiles that are otherwise difficult to access. NEV sample enrichment was validated using multiple methods, including TEM, NTA, and protein marker characterization (Fig. 1). Among the 128 miRNAs consistently detected across samples, brain-specific miRNAs, such as miR-134 and miR-128, were identified37,38. Furthermore, the target genes of these 128 miRNAs were significantly enriched in neuronal and brain tissues (Table S1), supporting the successful application of NEV-based approaches for analyzing brain-derived miRNA profiles in peripheral blood.
However, the specifications of L1CAM-based NEV isolation warrant discussion. Although L1CAM is widely used to isolate NEVs owing to its expression in neuronal tissues40,41,42, it is also expressed in peripheral nerve cells, Schwann cells, and other non-neuronal cell types, such as melanocytes, T cells, and B cells29,41. This study found an enrichment of genes expressed in both central and peripheral tissues, such as cardiac muscle fibers and sensory neurons, suggesting that the NEV samples might include EVs from L1CAM-expressing peripheral cells. Nevertheless, the detection of neuronal markers, such as ENO2 and MAPT, confirms the presence of brain-derived molecular profiles, particularly during disease onset, when brain tissue cannot be examined non-invasively (Fig. 1C).
Moreover, the miRNAs (miR-134 and miR-128) and protein markers (ENO2 and MAPT) detected in our study are highly expressed in neuronal tissues, according to previous reports43,44,45,46,47,48. However, they are not strictly brain-specific and can also be expressed in peripheral tissues. Therefore, detection of a single molecule is insufficient to establish a definitive neuronal origin. In this study, we combined multiple neuronal markers to estimate the neuronal origin, but we acknowledge that this approach has inherent limitations. Recently, ATP1A3 has been shown to provide greater neuronal specificity and enrichment efficiency than L1CAM or NCAM1 in EVs derived from the human brain, cerebrospinal fluid, and plasma35 and has been proposed as a promising alternative marker for NEV isolation. Furthermore, while some reports have raised concerns regarding soluble L1CAM contamination and the limited specificity of L1CAM+ EVs49,50, others, using single-EV analysis, have demonstrated the co-expression of neuronal markers within L1CAM+ EVs36. Taken together, these findings highlight the need for future studies to incorporate alternative markers, such as ATP1A3 or single-EV analyses, to further refine the evaluation of neuronal origin in NEV research.
Seventeen miRNAs were significantly differentially expressed in the ROS group compared with the CROS group, whereas two miRNAs were altered in the CS group compared with the CCS group (Fig. 2). Of the 18 miRNAs with significant changes in either comparison, 15 have been previously reported to be expressed in the central nervous system51, and 6 of these (hsa-miR-99a-5p, hsa-miR-193a-5p, hsa-miR-184, hsa-miR-375-3p, hsa-miR-423-3p, and hsa-miR-483-5p) have been associated with schizophrenia in postmortem brain studies52,53,54,55. In previous studies, four miRNAs (hsa-miR-122-5p, hsa-miR184, hsa-miR-342-5p, and hsa-miR-584-5p) were differentially expressed in the peripheral blood of patients with schizophrenia52,56,57. Among these, hsa-miR-184 was altered in both the recent-onset and chronic phases, suggesting its involvement throughout the disease trajectory. It has also been previously reported to be differentially expressed in postmortem brain and peripheral blood55,57.
Interestingly, most miRNAs that showed significant changes in the recent-onset phase did not exhibit any changes in the chronic phase. This finding highlights the potential importance of miRNAs in the early neurodevelopmental abnormalities that lead to schizophrenia. A previous study involving 221 patients with schizophrenia, mainly in the chronic phase, reported no miRNAs with expression changes greater than two-fold in NEVs, despite a larger sample size than that in the present study58. This consistency suggests that miRNA alterations are more pronounced during the recent-onset phase.
The differentially expressed miRNAs identified in this study revealed distinct functional roles during the recent-onset and chronic phases of schizophrenia. GO analysis highlighted the phase-specific and overlapping biological processes associated with these miRNAs, reflecting the dynamic changes in pathological mechanisms during disease progression.
During the recent-onset phase, miRNAs that were significantly altered in NEVs were associated with processes critical for neural circuit formation, such as synapse organization, axonogenesis, and dendrite development, particularly in glutamatergic and GABAergic neurons (Fig. 3A, Tables S6–S8). Across all three cell types examined, the miRNAs targeted genes involved in the regulation of neuronal projection development. These findings suggest that miRNA dysregulation during the onset phase may disrupt neurodevelopmental processes, particularly within 5 years of onsets, aligning with the neurodevelopmental hypothesis of schizophrenia59. Abnormalities in neural circuit formation contribute to the onset of schizophrenia and may represent key therapeutic targets for early intervention60,61.
In contrast, differentially expressed miRNAs in the chronic phase were associated with the regulation of synaptic plasticity and unfolded protein responses, such as the ER-nucleus signaling pathway and postsynaptic receptor regulation (Fig. 3B, Tables S9–S11). These processes are critical for maintaining synaptic homeostasis and cellular stress responses, particularly during the later stages of disease progression62,63. Abnormalities in synaptic plasticity have been extensively linked to the pathophysiology of schizophrenia, and targeting these processes may inform strategies for managing chronic symptoms64,65.
Interestingly, altered miRNAs in both phases have been implicated in synaptic transmission processes, including vesicle-mediated transport and trans-synaptic signaling. The dysregulation of these processes may underlie the persistent deficits in synaptic function observed throughout the course of the disease. Experimental evidence suggests that EVs regulate neural circuit formation, synaptic plasticity, and synaptic transmission through miRNA-mediated mechanisms, further supporting their role in schizophrenia pathogenesis30,66,67,68.
Among the recent-onset phase miRNAs, has-miR-143-3p exhibited the highest AUC as a diagnostic biomarker of ROS (Table 2) and was a promising candidate diagnostic biomarker. Furthermore, hsa-miR-143-3p stands out because of its prior association with high-risk children with schizophrenia69 and has been reported to be associated with schizophrenia70,71. This consistency across studies suggests that alterations in hsa-miR-143-3p expression may precede disease onset and potentially contribute to the development of schizophrenia. Additionally, hsa-miR-361-5p and hsa-miR-423-3p regulate key molecules, such as ITGB1 and RhoA72,73. ITGB1 plays a pivotal role in synapse formation by modulating cell-matrix interactions74, whereas RhoA is critical for neurodevelopmental processes, including dendritic spine density regulation, through Rho kinase signaling75,76. The dysregulation of these terms has been implicated in the pathogenesis of schizophrenia77,78,79.
For example, the downregulation of hsa-miR-361-5p in the recent-onset phase may lead to increased RhoA expression, potentially disrupting spine morphology and neural connectivity73,78. Furthermore, integrin-mediated synaptic changes associated with has-miR423-3p may contribute to the deficits in synaptic architecture observed in schizophrenia74,79. These findings suggest a mechanistic link between miRNA dysregulation and neurodevelopmental abnormalities during disease onset.
In the chronic phase, the association between miRNAs and unfolded protein response is particularly notable. Altered ER-nuclear signaling and synaptic receptor regulation suggest that miRNAs contribute to cellular stress responses and synaptic deficits, which may exacerbate chronic symptoms80,81. These processes align with previous findings of dysregulated protein homeostasis in the dorsolateral prefrontal cortex of patients with schizophrenia82. These insights highlight the potential of targeting miRNA-regulated pathways to ameliorate synaptic and cellular dysfunction in chronic schizophrenia.
This study has several limitations. First, the L1CAM-based enrichment method may not capture all NEVs, particularly those without L1CAM on their membranes38 and those that do not cross the blood-brain barrier (BBB), although NEVs have been reported to cross the BBB83,84. Further studies are required to explore NEV subpopulations and BBB permeability. Second, the small sample size limits the generalizability of the findings. Because this study was exploratory in nature, its results should be interpreted with caution. Replication and validation using larger independent cohorts are essential to confirm and extend these observations. Third, the CS group was administered high doses of antipsychotics. Although miRNAs associated with medication dosages were excluded from the functional analysis, it is possible that the miRNA expression profiles of the participants were affected by the medication dosages. Fourth, the CS and CCS groups in this study did not include female participants because of the availability of age-matched plasma samples. This sex imbalance limits the generalizability of our findings because sex differences are known to affect both schizophrenia pathophysiology and miRNA expression. Future studies should include a balanced number of male and female participants to validate and extend these results. Fifth, NTA and TEM characterizations were performed on whole EV preparations, rather than specifically on eluted L1CAM+ EVs. Although previous studies have successfully characterized L1CAM+ fractions by NTA and TEM35,36, the yield of L1CAM+ EVs in our plasma samples was insufficient for reliable measurements, and elution from antibody-coated beads may have altered vesicle morphology. Therefore, in this study, NTA and TEM were primarily used to confirm the presence, size distribution, and spherical morphology of the total EV population, from which the L1CAM+ subset was subsequently enriched. Although this approach ensures that L1CAM+ EVs are derived from bona fide EVs, direct morphological characterization was not performed in the present study. Similar strategies characterizing EVs at the total population level while focusing on specific downstream analyses of subpopulations have also been adopted in previous studies85. Furthermore, the L1CAM-based immunocapture method used here does not guarantee complete purity of L1CAM+ EVs, as non-vesicular or co-isolated molecules may also be present in the enriched fraction. This represents a major limitation of the present study, and future improvements in immunocapture yield and single-EV validation will be necessary to achieve more precise molecular characterization. Finally, the mechanisms underlying NEV uptake by recipient cells remain unclear. While some studies suggest selectivity, others indicate variability based on EV size and density86,87,88. Elucidating these mechanisms may enhance our understanding of NEV-mediated intercellular communication.
To our knowledge, this is the first study to use NEVs to investigate miRNA expression shortly after the onset of schizophrenia. Our findings highlight possible miRNA-related pathological mechanisms in the recent onset and chronic phases of schizophrenia, emphasizing the importance of neurodevelopmental changes in schizophrenia. Despite these limitations, this study highlights the utility of NEVs as a minimally invasive tool for accessing brain-derived molecular profiles. The identified miRNAs, particularly those that were differentially expressed during the recent-onset phase, show promise as diagnostic biomarkers and provide new insights into the pathogenesis of schizophrenia. Further research is required to validate these findings and explore their therapeutic implications.
Methods
Study design and participants
The participants included 23 healthy controls and 22 patients with schizophrenia, divided into two groups: ROS and CS. Schizophrenia was diagnosed by at least two experienced psychiatrists based on the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) or 5th edition (DSM-V) guidelines. Among the 22 patients, 14 were classified as having ROS (within 5 years of onset), and 8 were classified as having CS (15 years or more after onset).
All the patients with schizophrenia received therapeutic medication. In the ROS group, 11 of 14 patients were taking antipsychotics (three patients not taking antipsychotics were taking anxiolytics or antidepressants), four were taking anxiolytics, one was taking antidepressants, and one was using benzodiazepine receptor agonist hypnotics. In the CS group, all eight patients were taking antipsychotics, four were taking anxiolytics, and seven were using benzodiazepine receptor agonist hypnotics, with none taking antidepressants. Medication dosages were standardized as chlorpromazine equivalents for antipsychotics, diazepam equivalents for anxiolytics, imipramine equivalents for antidepressants, and nitrazepam equivalents for benzodiazepine receptor agonist hypnotics.
At the time of blood collection, inpatient or outpatient status was recorded, and symptom severity was assumed using the PANSS. In the ROS group, nine patients were outpatients and five were inpatients; in the CS group, two patients were outpatients and six were inpatients. PANSS scores were available for 11 patients in the ROS group and all eight patients in the CS group. The mean total PANSS score in the ROS group was 38.2 ± 8.8, corresponding to mild symptom severity and indicating that these patients were not acutely psychotic at the time of blood sampling. Clinical evaluation by psychiatrists at the same time also confirmed that none of the patients were in an acute psychotic state. By contrast, the mean total PANSS score in the CS group was 106.5 ± 12.4, reflecting persistent and more severe psychopathology typical of chronic schizophrenia. Importantly, none of the patients in either group were administered clozapine. These clinical details are summarized in Table 1.
A group of 15 controls, matched for age and sex to the ROS group (CROS), and a group of eight controls, matched for age and sex to the CS group (CCS), were selected as healthy controls from a pooled sample of participants. In addition, smoking and drinking habits were assessed in a subset of the participants.
This study was approved by the ethics committees of all the participating institutions (Tokyo Metropolitan Matsuzawa Hospital and Tokyo Metropolitan Institute of Medical Science, approval numbers 17-16, 20-17, and 23-12). Written informed consent was obtained from all the patients, and the study adhered to the principles of the Declaration of Helsinki.
Sampling and EV isolation
Blood samples were collected between 9:00 AM and 6:00 PM on designated collection days. Plasma was obtained by centrifuging blood samples at 1200 × g for 5 min at 4 °C. For thrombin treatment, 0.5 mL of plasma was incubated with 4 µL of thrombin (System Biosciences, Mountain View, CA, USA) at room temperature for 5 min, followed by centrifugation at 9000 × g for 5 min. The supernatants were collected for EV isolation.
EVs were isolated using 63 µL of ExoQuick (System Biosciences), followed by incubation at 4 °C for 2 h and centrifugation at 1500 × g for 30 min. The resulting pellet containing EVs was resuspended in 0.5 mL of phosphate-buffered saline containing a protease and phosphatase inhibitor cocktail (Roche, Indianapolis, IN, USA).
Nanoparticle characterization and transmission electron microscopy analysis
NTA was performed at FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan) to assess EV size and concentration. The EV suspensions were diluted to a particle concentration of approximately 108–109 particles/mL using Milli-Q water, and the size distributions were measured using NanoSight LM10 (Malvern Instruments, Malvern, UK). TEM was performed at Hanaichi Ultrastructure Research Institute (Okazaki, Japan) using negatively stained EV samples. Briefly, a drop of the sample was placed onto a 200-mesh formvar/carbon-coated copper grid and allowed to settle for 3 min at room temperature. The grids were washed with distilled water and 20 μL of an aqueous solution of uranyl acetate (2% w/v), then applied for 10 s. The samples were then analyzed using a JEOL JEM1400Flash TEM (Jeol Ltd., Tokyo, Japan) operating at 100 KV.
Enrichment and characterization of NEVs
NEVs were enriched using the Exo-Flow Beads and Buffers Kit (System Biosciences). Thirty-five microliters of streptavidin magnetic beads were conjugated to 4 µg biotinylated anti-human CD171 (L1CAM) antibody (clone 5G3, cat. 13-1719-82, RRID AB-2043813; eBiosciences, San Diego, CA, USA) through incubation at 4 °C for 2 h. The antibody-conjugated beads were then incubated with the 500 µl of EV suspension overnight at 4 °C to capture NEVs. Beads were washed three times with 500 µl of wash buffer from the kit. NEVs were eluted using 700 µl QIAzol Lysis Reagent (Qiagen, Hilden, Germany) and characterized using the Exo-Check Exosome Antibody Array Neuro Mini (System Biosciences), following the manufacturer’s instructions.
These procedures, including NTA, TEM, and immunoblot/antibody array analyses of EV markers, were conducted in accordance with the MISEV 2023 guidelines34.
miRNA extraction and sequencing
RNA was extracted from NEVs using the miRNEASY Mini Kit (Qiagen) and stored at −80 °C. RNA quality was assessed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), confirming small RNA enrichment (<200 nt). Library preparation was performed using the NEBNext Small RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA), and sequencing was conducted on a NovaSeq 6000 platform (Illumina, San Diego, CA, USA) with a 101-bp single-end read at the Research Institute for Microbial Diseases, Osaka University (Osaka, Japan). Sequencing generated 1,197,843,508 reads (an average of 26,618,744 reads per donor), indicating saturation and reliable quantification. Data quality was assessed using FastQC v0.11.989, and adapter sequences were trimmed using Cutadapt v4.090. Alignment to the human reference genome GRCh38 release 11091 was performed using Bowtie2 v2.5.192, and BAM files were constructed using SAMtools v1.1893.
miRNA statistical analysis
The miRNA expression was normalized using the relative log expression method94 option in the Bioconductor/edgeR package (v3.42.4)95. Differential expression analysis was conducted using the likelihood ratio test, comparing the ROS group to the CROS group and the CS group to the CCS group. The full model included disease status, age, and sex as explanatory variables, whereas the reduced model included only age and sex. P values were adjusted for multiple comparisons using the Benjamini–Hochberg FDR procedure.
Multiple regression analysis was conducted to explore the associations between miRNA expression and clinical or behavioral variables, such as medication dose, smoking, and alcohol consumption. Some participants with no data on clinical or behavioral variables were excluded from this analysis, leaving 32 participants. For each miRNA, the regression model used log-transformed normalized expression levels as the dependent variable and disease status, age, sex, and clinical or behavioral variables as independent variables. miRNAs with p values below 0.05 for any independent variable were considered significantly associated with that variable.
Diagnostic biomarker evaluation
Statistical analyses were performed using R version 4.3.1. Univariate logistic regression analyses were used to identify the miRNAs with the strongest discriminatory power between the ROS and CROS groups using the glm function in R. Among the 15 miRNAs that showed significant differential expression and were not associated with medication dosage, each was individually evaluated as an explanatory variable for predicting ROS status. miRNA expression levels were log-transformed prior to analysis. The AUC and 95% CI were calculated to quantify diagnostic performance using the pROC R package. The sensitivity, specificity, accuracy, and threshold were evaluated using 5-fold cross-validation to ensure robustness and reduce overfitting. P values were adjusted for multiple comparisons using the Benjamini–Hochberg FDR procedure.
Target genes’ functional analysis
To further understand the functional implications of the differentially expressed miRNAs, predicted target genes were identified using miRWalk version 396. The predicted targets were subjected to tissue-specific enrichment analysis using EnrichR97, focusing on the ARCHS4 tissue database98.
Functional analysis of the target genes was performed using over-representation analysis on the web-based gene set analysis toolkit 202499, applying GO analysis of the biological process (“noRedundant” option was applied). This analysis focused on genes expressed in glutamatergic neurons, GABAergic neurons, and astrocytes, based on single-nucleus sequencing data from Brodmann area 46 in postmortem brain samples39. Genes were defined as being expressed in a cell type if they exhibited at least one unique molecular identifier count across all donors, excluding outlier donors. The p values from the tissue-specific enrichment and GO analyses were adjusted using the Benjamini–Hochberg FDR procedure to account for multiple comparisons.
Data availability
The data that support the findings of this study are available upon reasonable request from the corresponding author, K.T.
References
Bartel, D. M. MicroRNAs. Cell 173, 20–51 (2018).
Aksoy-Aksel, A. & Zampa, F. G. MicroRNAs and synaptic plasticity—a mutual relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130515 (2014).
Ziats, M. N. & Rennert, O. M. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 19, 848–852 (2014).
Moreau, M. P., Bruse, S. E., David-Rus, R., Buyske, S. & Brzustowicz, L. M. Altered MicroRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 69, 188–193 (2011).
Warnica, W. et al. Copy number variable micrornas in schizophrenia and their neurodevelopmental gene targets. Biol. Psychiatry 77, 158–166 (2015).
Hu, Z. et al. Temporal dynamics of miRNAs in human DLPFC and its association with miRNA dysregulation in schizophrenia. Transl. Psychiatry 9, 196 (2019).
Forstner, A. J., Degenhardt, F., Schratt, G. & Nöthen, M. M. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front. Mol. Neurosci. 5, 47 (2013).
Solmi, M. et al. Age at onset of mental disorders worldwide: large-scale meta-analysis of 192 epidemiological studies. Mol. Psychiatry 27, 281–295 (2022).
Hafner, H., Maurer, K., Loffler, W. & Riecher-Rossler, A. The influence of age and sex on the onset of early course of schizophrenia. Br. J. Psychiatry 162, 80–86 (1993).
Durston, S. et al. Anatomical MRI of the developing human brain: What have we learned?. J. Am. Acad. Child Adolesc. Psychiatry 40, 1012–1020 (2001).
Casey, B. J., Jones, R. M. & Hare, T. A. The adolescent brain. Ann. N. Y Acad. Sci. 1124, 111–126 (2008).
Paus, T. Mapping brain maturation and cognitive development during adolescence. Trends Cogn. Sci. 9, 60–68 (2005).
Giedd, J. N. et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat. Neurosci. 2, 861–863 (1999).
Marenco, S. & Weinberger, D. R. The neurodevelopmental hypothesis of schizophrenia: following a trail of evidence from cradle to grave. Dev. Psychopathol. 12, 501–527 (2000).
Birnbaum, R. & Weinberger, D. R. The genesis of schizophrenia: an origin story. Am. J. Psychiatry 181, 482–492 (2024).
Paus, T., Keshavan, M. & Giedd, J. N. Why do many psychiatric disorders emerge during adolescence?. Nat. Rev. Neurosci. 9, 947–957 (2008).
McGorry, P. D. et al. The Lancet Psychiatry Commission on youth mental health. Lancet Psychiatry 11, 731774 (2024).
Thomas, K. T. & Zakharenko, S. S. MicroRNAs in the onset of Schizophrenia. Cells 6, 10 (2021).
Rahbarghazi, R. et al. Tumorderived extracellular vesicles: reliable tools for cancer diagnosis and clinical applications. Cell Commun. Signal. 17, 73 (2019).
Badhwar, A. P. & Haqqani, A. S. Biomarker potential of brain-secreted extracellular vesicles in blood in Alzheimer’s disease. Alzheimers Dement. 12, 12001 (2020).
Wang, X., Yang, H., Liu, C. & Liu, K. A new diagnostic tool for brain disorders: extracellular vesicles derived from neuron, astrocyte, and oligodendrocyte. Front. Mol. Neurosci. 16, 1194210 (2023).
Heinzelman, P., Bilousova, T., Campagna, J. & John, V. Nanoscale extracellular vesicle analysis in Alzheimer’s disease diagnosis and therapy. Int. J. Alzheimers Dis. 2016, 8053139 (2016).
Pegtel, D. M. & Gould, S. J. Exosomes. Annu. Rev. Biochem 59, 48 (2019).
Murillo, O. D. et al. exRNA Atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell 177, 463–477 (2019).
Merchant, M. L., Rood, I. M., Deegens, J. J. & Klein, J. B. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 13, 731–749 (2017).
Todorova, D., Simoncini, S., Lacroix, R., Sabatier, F. & Dignat-George, F. Extracellular vesicles in angiogenesis. Cir. Res. 12, 1658–1673 (2017).
Rufino-Ramos, D. et al. Extracellular vesicles: novel promising delivery systems for therapy of brain diseases. J. Control. Release 262, 247–258 (2017).
Guo, C. et al. Circular RNAs in extracellular vesicles: promising candidate biomarkers for schizophrenia. Front. Genet. 6, 997322 (2023).
Gomes, D. E. & Witwer, K. W. L1CAM-associated extracellular vesicles: a systematic review of nomenclature, sources, separation, and characterization. J. Extracell. Biol. 2, 95 (2023).
Goldie, B. J. et al. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 42, 9195–9208 (2014).
Raghavan, V., Bhomia, M., Torres, I., Jain, S. & Wang, K. K. Hypothesis: exosomal microRNAs as potential biomarkers for schizophrenia. Med. Hypotheses 103, 21–25 (2017).
van den Berg, M. J. et al. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 185, 101732 (2020).
Mustapic, M. et al. Plasma extracellular vesicles enriched for neuronal origin: a potential window into brain pathologic processes. Front. Neurosci. 11, 278 (2017).
Welsh, J. A. et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 13, e12451 (2024).
You, Y. et al. ATP1A3 as a target for isolating neuron specific extracellular vesicles from human brain and biofluids. Sci. Adv. 9, eadi3647 (2023).
Nogueras Ortiz, C. J. etal. Single extracellular vesicle (EV) analyses validate the use of L1 Cell Adhesion Molecule (L1CAM) as a reliable biomarker of neuron derived EVs. J. Extracell. Vesicles 13, e12459 (2024).
Schratt, G. M. et al. A brainspecific microRNA regulates dendritic spine development. Nature 439, 283–289 (2006).
Lagos-Quintana, M. et al. Identification of tissue-specific MicroRNAs from mouse. Curr. Biol. 12, 735–739 (2002).
Ling, E. et al. A concerted neuron–astrocyte program declines in ageing and schizophrenia. Nature 627, 604611 (2024).
Fiandaca, M. S. et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement. 11, 600–607 (2005).
Bravo-Miana, R. C., Arizaga-Echebarria, J. K. & Otaegui, D. Central nervous system-derived extracellular vesicles: the next generation of neural circulating biomarkers?. Transl. Neurodegener. 19, 32 (2024).
Taha, H. B. & Ati, S. S. Evaluation of α-synuclein in CNS-originating extracellular vesicles for Parkinsonian disorders: a systematic review and meta-analysis. CNS Neurosci. Ther. 29, 3741–3755 (2023).
Wu, Y. H. et al. miR-134 modulates the proliferation of human cardiomyocyte progenitor cells by targeting Meis2. Int. J. Mol. Sci. 16, 25199–25213 (2015).
Li, L., He, X., Liu, M., Yun, L. & Cong, B. Diagnostic value of cardiac miR-126-5p, miR-134-5p, and miR-499a-5p in coronary artery disease-induced sudden cardiac death. Front. Cardiovasc. Med. 9, 944317 (2022).
Hooten, N. N. et al. microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE 5, e10724 (2010).
Zheng, Y. et al. Insulin-like growth factor 1-induced enolase 2 deacetylation by HDAC3 promotes metastasis of pancreatic cancer. Signal Transduct. Target. Ther. 5, 53 (2020).
Gu, Y., Oyama, F. & Ihara, Y. Tau is widely expressed in rat tissues. J. Neurochem 67, 1235–1244 (1996).
Rouzier, R. et al. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc. Natl. Acad. Sci. USA 102, 8315–8320 (2005).
Norman, M. et al. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat. Methods 18, 631–634 (2021).
Kadam, V. et al. Most L1CAM is not associated with extracellular vesicles in human biofluids and iPSC-derived neurons. Mol. Neurobiol. 62, 10427–10442 (2025).
Panwar, B., Omenn, G. S. & Guan, Y. miRmine: a database of human miRNA expression profiles. Bioinformatics 33, 1554–1560 (2017).
Gardiner, E. et al. Imprinted DLK1DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol. Psychiatry 17, 827–840 (2012).
Rey, R., Suaud-Chagny, M. F., Dorey, J. M., Teyssier, J. R. & d’Amato, T. Widespread transcriptional disruption of the microRNA biogenesis machinery in brain and peripheral tissues of individuals with schizophrenia. Transl. Psychiatry 10, 376 (2020).
Santarelli, D. M., Beveridge, N. J., Tooney, P. A. & Cairns, M. J. Upregulation of Dicer and MicroRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 69, 180–187 (2011).
Beveridge, N. J., Gardiner, E., Carroll, A. P., Tooney, P. A. & Cairns, M. J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 15, 1176–1189 (2010).
Wei, H. et al. Detection of circulating miRNA levels in schizophrenia. Am. J. Psychiatry 172, 1141–1147 (2015).
Du, Y. et al. Genome-wide, integrative analysis implicates exosome-derived MicroRNA dysregulation in schizophrenia. Schizophr. Bull. 45, 1257–1266 (2019).
Barnett, M. M. et al. MiRNA cargo in circulating vesicles from neurons is altered in individuals with schizophrenia and associated with severe disease. Sci. Adv. 9, 4386 (2009).
Waltereit, R., Banaschewski, T., Meyer-Lindenberg, A. & Poustka, L. Interaction of neurodevelopmental pathways and synaptic plasticity in mental retardation, autism spectrum disorder and schizophrenia: implications for psychiatry. World J. Biol. Psychiatry 15, 507–516 (2014).
Lee, H. et al. Early cognitive experience prevents adult deficits in a neurodevelopmental schizophrenia model. Neuron 7, 714–724 (2012).
Mikanmaa, E. et al. Towards a neurodynamical understanding of the prodrome in schizophrenia. Neuroimage 190, 144–153 (2019).
Nevell, L. et al. Elevated systemic expression of ER stress related genes is associated with stress-related mental disorders in the Detroit Neighborhood Health Study. Psychoneuroendocrinology 43, 62–70 (2014).
Prabakaran, S. et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 9, 684–697 (2004).
Dudzik, P., Lustyk, K. & Pytka, K. Beyond dopamine: novel strategies for schizophrenia treatment. Med. Res. Rev. 44, 2307–2330 (2024).
Forsyth, J. K. & Lewis, D. A. Mapping the consequences of impaired synaptic plasticity in schizophrenia through development: an integrative model for diverse clinical features. Trends Cogn. Sci. 21, 760–778 (2017).
Zaki, M. B. et al. The potential role of miRNAs in the pathogenesis of schizophrenia—a focus on signaling pathways interplay. Pathol. Res Pract. 254, 155102 (2024).
Lafourcade, C. et al. MiRNAs in astrocytederived exosomes as possible mediators of neuronal plasticity. J. Exp. Neurosci. 8, 1–9 (2016).
Chamakioti, M., Chrousos, G. P., Kassi, E., Vlachakis, D. & Yapijakis, C. Stress-related roles of exosomes and exosomal miRNAs in common neuropsychiatric disorders. Int. J. Mol. Sci. 29, 8256 (2024).
Tomita, Y. et al. Urinary exosomal microRNAs as predictive biomarkers for persistent psychotic-like experiences. Schizophrenia 9, 14 (2023).
Sargazi, S. et al. Functional variants of miR-143 are associated with schizophrenia susceptibility: a preliminary population-based study and bioinformatics analysis. Biochem. Genet. 60, 868–881 (2022).
Waddington, J. L., Wang, X. & Zhen, X. ‘Whole-Body’ perspectives of schizophrenia and related psychotic illness: miRNA-143 as an exemplary molecule implicated across multi-system dysfunctions. Biomolecules 20, 1185 (2024).
Karginov, F. V. & Hannon, G. J. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 27, 1624–1632 (2013).
Kishore, S. et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559–567 (2011).
Wu, X. & Reddy, D. S. Integrins as receptor targets for neurological disorders. Pharmacol. Ther. 134, 68–81 (2012).
Li, Z., Aizenman, C. D. & Cline, H. T. Regulation of Rho GTPases by crosstalk and neuronal activity in vivo. Neuron 33, 741–750 (2022).
Huang, G. H., Sun, Z. L., Li, H. J. & Feng, D. F. Rho GTPase-activating proteins: regulators of Rho GTPase activity in neuronal development and CNS diseases. Mol. Cell. Neurosci. 80, 18–31 (2017).
Hill, J. J., Hashimoto, T. & Lewis, D. A. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry 11, 557–566 (2006).
Tanaka, R. & Yamada, K. Genomic and reverse translational analysis discloses a role for small GTPase RhoA signaling in the pathogenesis of schizophrenia: Rho-kinase as a novel drug target. Int. J. Mol. Sci. 24, 15623 (2023).
Berretta, S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology 62, 1584–1597 (2022).
Gonzalez-Liencres, C. et al. Oxidative stress in schizophrenia: a case-control study on the effects on social cognition and neurocognition. BMC Psychiatry 14, 268 (2014).
Cuenod, M. et al. Caught in vicious circles: a perspective on dynamic feed-forward loops driving oxidative stress in schizophrenia. Mol. Psychiatry 27, 1886–1897 (2022).
Kim, P., Scott, M. R. & Meador-Woodruff, J. H. Dysregulation of the unfolded protein response (UPR) in the dorsolateral prefrontal cortex in elderly patients with schizophrenia. Mol. Psychiatry 26, 1321–1331 (2021).
Khatami, S. H. et al. Exosomes: promising delivery tools for overcoming blood-brain barrier and glioblastoma therapy. Mol. Neurobiol. 60, 4659–4678 (2023).
Banks, W. A. et al. Transport of extracellular vesicles across the blood-brain barrier: brain pharmacokinetics and effects of inflammation. Int. J. Mol. Sci. 21, 1–21 (2020).
Chou, S. Y. et al. Altered insulin receptor substrate 1 phosphorylation in blood neuron-derived extracellular vesicles from patients with Parkinson’s disease. Front. Cell Dev. Biol. 8, 564641 (2020).
Venturini, A. et al. Exosomes from astrocyte processes: signaling to neurons. Front. Pharmacol. 10, 1452 (2019).
Huo, L., Du, X., Li, X., Liu, S. & Xu, Y. The emerging role of neural cell-derived exosomes in intercellular communication in health and neurodegenerative diseases. Front. Neurosci. 15, 738442 (2021).
Abdelsalam, M., Ahmed, M., Osaid, Z., Hamoudi, R. & Harati, R. Insights into exosome transport through the blood–brain barrier and the potential therapeutical applications in brain diseases. Pharmaceuticals 16, 571 (2023).
Andrews, S., Krueger, F., Seconds-Pichon, A., Biggins, F. & Wingett, S. Babraham bioinformatics-FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011).
Perez, G. et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res. 53, D1243–D1249 (2025).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Sticht, C., De La Torre, C., Parveen, A. & Gretz, N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE 13, e0206239 (2018).
Chen, E. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128 (2013).
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Elizarraras, J. M. et al. WebGestalt 2024: faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 52, W415–W421 (2024).
Acknowledgements
The authors express their sincere gratitude to Eriko Makiyama for her assistance. The authors would also like to thank all the participants and their families.
Author information
Authors and Affiliations
Contributions
Y.T. conceived and designed the study, conducted the experiments, performed RNA-seq data analyses, and drafted the initial manuscript. K.T. contributed to study design, performed clinical analyses of the patient cohort, and played a major role in revising the manuscript. K.T., M.A., and M.I. supervised the study. M.M., K.S., and M.I. collected clinical samples and information. H.K. provided support for RNA-seq data analysis. All other authors reviewed the final manuscript. This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 23K27535 to M.A., 24H00666, 25K02595 to K.T., and 24K18719 to K.S.) and a Grant-in-Aid for JSPS Fellows (grant number JP22J22871 to Y.T.). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
Y.T., K.T., M.M., K.S., H.K., M.I., and M.A. received grants from the Japan Society for the Promotion of Science. The authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tomita, Y., Toriumi, K., Miyashita, M. et al. Distinct microRNA profiles in neuron-derived extracellular vesicles between recent-onset and chronic-phase schizophrenia. Schizophr 11, 158 (2025). https://doi.org/10.1038/s41537-025-00706-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41537-025-00706-x