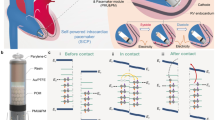
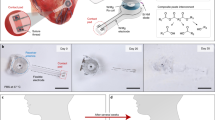
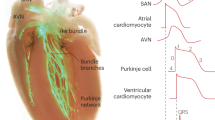
Abstract
Bio-pacemakers offer a potential alternative to electronic devices, yet their stable implementation at cellular and tissue levels remains unresolved. In this computational study, we aimed to investigate possible effects of the electrotonic interaction between cardiac cells and the spatial distribution of the bio-pacemaker on the initiation and conduction of cardiac pacemaking action potentials to surrounding quiescent cardiac tissues. Simulation results demonstrated that (i) a combination of weak gap junctional electrical coupling among PMs; and (ii) rectified coupling arising from heterotypic gap junction expressions between the PM and ventricle yielded the best stable and robust bio-pacemaker for pacing and driving surrounding ventricular tissue. Furthermore, Isolated or septal placement improved ventricular pacing efficacy. This study adopts a digital health approach, providing an important theoretical foundation for the simulation of new clinical therapies.
Similar content being viewed by others
Data availability
This study is based on computational simulations, and all data can be derived analytically from the equations presented. The relevant mathematical formulations and derivation steps are provided in the Supplementary Materials.
Code availability
The computational code used in this study was developed for research purposes and is available from the corresponding author upon reasonable request.
References
Cingolani, E., Goldhaber, J. I. & Marban, E. Next-generation pacemakers: from small devices to biological pacemakers. Nat. Rev. Cardiol. 15, 139–150 (2018).
Cho, H. C. & Marban, E. Biological therapies for cardiac arrhythmias: can genes and cells replace drugs and devices?. Circ. Res. 106, 674–685 (2010).
Rosen, M. R., Brink, P. R., Cohen, I. S. & Robinson, R. B. Genes, stem cells and biological pacemakers. Cardiovasc. Res. 64, 12–23 (2004).
Shlapakova, I. N. et al. Biological pacemakers in canines exhibit positive chronotropic response to emotional arousal. Heart Rhythm 7, 1835–1840 (2010).
Grijalva, S. I. et al. Engineered cardiac pacemaker nodes created by TBX18 gene transfer overcome source-sink mismatch. Adv. Sci. 6, 1901099 (2019).
Kapoor, N., Liang, W., Marban, E. & Cho, H. C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 31, 54–62 (2013).
Gorabi, A. M. et al. TBX18 transcription factor overexpression in human-induced pluripotent stem cells increases their differentiation into pacemaker-like cells. J. Cell. Physiol. 234, 1534–1546 (2019).
Hu, Y. et al. Genetically modified porcine mesenchymal stem cells by lentiviral Tbx18 Create a biological pacemaker. Stem Cells Int. 2019, 1–9 (2019).
Li, Y., Wang, K., Li, Q., Hancox, J. C. & Zhang, H. Reciprocal interaction between IK1 and If in biological pacemakers: a simulation study. PLoS Comput. Biol. 17, e1008177 (2021).
Kurata, Y., Matsuda, H., Hisatome, I. & Shibamoto, T. Effects of pacemaker currents on creation and modulation of human ventricular pacemaker: theoretical study with application to biological pacemaker engineering. Am. J. Physiol. Heart Circ. Physiol. 292, H701–H718 (2007).
Silva, J. & Rudy, Y. Mechanism of pacemaking in I(K1)-downregulated myocytes. Circ. Res. 92, 261–263 (2003).
Plotnikov, A. N. et al. Xenografted adult human mesenchymal stem cells provide a platform for sustained biological pacemaker function in canine heart. Circulation 116, 706–713 (2007).
Hu, Y. F., Dawkins, J. F., Cho, H. C., Marban, E. & Cingolani, E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 6, 1–10 (2014).
Jongsma, H. J. Diversity of gap junctional proteins: does it play a role in cardiac excitation?. J. Cardiovasc. Electrophysiol. 11, 228–230 (2000).
Inada, S. et al. Importance of gradients in membrane properties and electrical coupling in sinoatrial node pacing. PLoS ONE 9, e94565 (2014).
Rackauskas, M. et al. Gating properties of heterotypic gap junction channels formed of connexins 40, 43, and 45. Biophys. J. 92, 1952–1965 (2007).
Valiunas, V., Weingart, R. & Brink, P. R. Formation of heterotypic gap junction channels by connexins 40 and 43. Circ. Res. 86, E42–E49 (2000).
Jansen, J. A., van Veen, T. A., de Bakker, J. M. & van Rijen, H. V. Cardiac connexins and impulse propagation. J. Mol. Cell. Cardiol. 48, 76–82 (2010).
Honjo, H. et al. Heterogeneous expression of connexins in rabbit sinoatrial node cells: correlation between connexin isotype and cell size. Cardiovasc. Res. 53, 89–96 (2002).
Coppen, S. R. et al. Connexin45, a major connexin of the rabbit sinoatrial node, is co-expressed with connexin43 in a restricted zone at the nodal-crista terminalis border. J. Histochem. Cytochem. 47, 907–918 (1999).
Mangion, K., Gao, H., Husmeier, D., Luo, X. & Berry, C. Advances in computational modelling for personalised medicine after myocardial infarction. Heart 104, 550–557 (2018).
ten Tusscher, K. H. & Panfilov, A. V. Alternans and spiral breakup in a human ventricular tissue model. Am. J. Physiol. Heart Circ. Physiol. 291, H1088–H1100 (2006).
Severs, N. J., Bruce, A. F., Dupont, E. & Rothery, S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res. 80, 9–19 (2008).
Valiunas, V. et al. Coupling an HCN2-expressing cell to a myocyte creates a two-cell pacing unit. J. Physiol. 587, 5211–5226 (2009).
Li, Y., Ma, L., Li, Q., Zhang, H. & Wang K. (eds) Effect of cell coupling between pacemaker cells on the biological pacemaker in cardiac tissue model. In Proc. 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (IEEE, 2022).
Moreno, A. P. Biophysical properties of homomeric and heteromultimeric channels formed by cardiac connexins. Cardiovasc. Res. 62, 276–286 (2004).
Ye, W. G. et al. Junctional delay, frequency, and direction-dependent uncoupling of human heterotypic Cx45/Cx43 gap junction channels. J. Mol. Cell. Cardiol. 111, 17–26 (2017).
Bukauskas, F. F., Angele, A. B., Verselis, V. K. & Bennett, M. V. Coupling asymmetry of heterotypic connexin 45/connexin 43-EGFP gap junctions: properties of fast and slow gating mechanisms. Proc. Natl. Acad. Sci. USA 99, 7113–7118 (2002).
Urthaler, F., Katholi, C. R., Macy, J. R. J. & James, T. N. Electrophysiological and mathematical characteristics of the escape rhythm during complete AV block. Cardiovasc. Res. 8, 173–186 (1974).
Plotnikov, A. N. et al. Biological pacemaker implanted in canine left bundle branch provides ventricular escape rhythms that have physiologically acceptable rates. Circulation 109, 506–512 (2004).
Li, N. et al. Redundant and diverse intranodal pacemakers and conduction pathways protect the human sinoatrial node from failure. Sci. Transl. Med. 9, 1–11 (2017).
Fedorov, V. V., Glukhov, A. V. & Chang, R. Conduction barriers and pathways of the sinoatrial pacemaker complex: their role in normal rhythm and atrial arrhythmias. Am. J. Physiol. Heart Circ. Physiol. 302, H1773–H1783 (2012).
Zhang, Y., Liu, S. Y., Li, J. & Wang, K. Q. Effect of electrical coupling at the border on the 1D biological pacemaker—a simulation study. Prog. Biochem. Biophys. 47, 448–456 (2020).
Unudurthi, S. D., Wolf, R. M. & Hund, T. J. Role of sinoatrial node architecture in maintaining a balanced source-sink relationship and synchronous cardiac pacemaking. Front. Physiol. 5, 1–7 (2014).
de Jong, M. A. & Merks, R. M. In silico model suggests that interdigitation promotes robust activation of atrial cells by pacemaker cells. Preprint at bioRxiv https://doi.org/10.1101/2024.05.15.594103 (2024).
de Cock, C. C., Giudici, M. C. & Twisk, J. W. Comparison of the haemodynamic effects of right ventricular outflow-tract pacing with right ventricular apex pacing: a quantitative review. Europace 5, 275–278 (2003).
Vardas, P. E., Simantirakis, E. N. & Kanoupakis, E. M. New developments in cardiac pacemakers. Circulation 127, 2343–2350 (2013).
Sweeney, M. O. & Prinzen, F. W. A new paradigm for physiologic ventricular pacing. J. Am. Coll. Cardiol. 47, 282–288 (2006).
Clayton, R. H. et al. Models of cardiac tissue electrophysiology: progress, challenges and open questions. Prog. Biophys. Mol. Biol. 104, 22–48 (2011).
Li, Y. et al. In silico mechanisms of arsenic trioxide-induced cardiotoxicity. Front. Physiol. 13, 1004605 (2022).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) under Grant No. 62133009 and No. 61702026, and the Open Project Program of State Key Laboratory of Virtual Reality Technology and Systems, Beihang University, under Grant No. VRLAB2024A05.
Author information
Authors and Affiliations
Contributions
Conceptualisation: Qince Li, Henggui Zhang; Data curation: Yacong Li, Dong Sui; Formal analysis: Yacong Li, Xiangyun Bai; Funding acquisition: Yacong Li, Dong Sui, Qince Li; Investigation: Yacong Li, Dong Sui, Deyan Yang; Methodology: Yacong Li, Jun Liu, Xiangyun Bai, Qince Li; Project administration: Lei Ma, Kuanquan Wang, Henggui Zhang; Resources: Yacong Li, Qince Li, Lei Ma; Software: Yacong Li, Xiangyun Bai, Dong Sui; Supervision: Kuanquan Wang, Henggui Zhang; Validation: Yacong Li, Deyan Yang; Visualisation: Yacong Li, Dong Sui; Writing – original draft: Yacong Li, Jun Liu, Xiangyun Bai, Dong Sui; Writing – review & editing: Qince Li, Lei Ma, Kuanquan Wang, Henggui Zhang.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Liu, J., Bai, X. et al. Mechanisms of rectified gap junctional coupling enhancing pacemaking activity of biologically engineered pacemaker cells. npj Syst Biol Appl (2026). https://doi.org/10.1038/s41540-025-00646-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41540-025-00646-3