Abstract
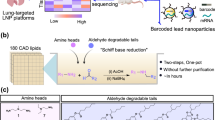
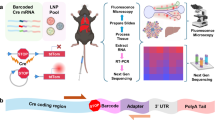
Developing safe and effective nanoparticles for the delivery of messenger RNA (mRNA) is slow and expensive, partly due to the lack of predictive power of in vitro screening methods and the low-throughput nature of in vivo screening. While DNA barcoding and batch analysis present methods for increasing in vivo screening throughput, they can also result in incomplete or misleading measures of efficacy. Here, we describe a high-throughput and accurate method for the screening of pooled nanoparticle formulations within the same animal. The method uses liquid chromatography with tandem mass spectrometry to detect peptide barcodes translated from mRNAs in nanoparticle-transfected cells. We show the method’s applicability by evaluating a library of over 400 nanoparticle formulations with 384 unique ionizable lipids using only nine mice to optimize the formulation of a biodegradable lipid nanoparticle for mRNA delivery to the liver. Barcoding lipid nanoparticles with peptide-encoding mRNAs may facilitate the rapid development of nanoparticles for mRNA delivery to specific cells and tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. All data generated or analysed during the study are available from the corresponding author on reasonable request.
References
Yin, H. et al. Non-viral vectors for gene-based therapy. Nat Rev. Genet. 15, 541–555 (2014).
Kauffman, K. J., Webber, M. J. & Anderson, D. G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release 240, 227–234 (2016).
Li, B., Zhang, X. & Dong, Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11, e1530 (2019).
Kowalski, P. S., Rudra, A., Miao, L. & Anderson, D. G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 27, 710–728 (2019).
Rhym, L. H. & Anderson, D. G. Nanoscale delivery platforms for RNA therapeutics: challenges and the current state of the art. Med 3, 167–187 (2022).
DeRosa, F. et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 23, 699–707 (2016).
Pardi, N. et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543, 248–251 (2017).
Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019).
Yin, H. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 (2016).
Miller, J. B. et al. Non-Viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. https://doi.org/10.1002/anie.201610209 (2017).
Yin, H. et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179–1187 (2017).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Whitehead, K. A. et al. In vitro - in vivo translation of lipid nanoparticles for hepatocellular siRNA delivery. ACS Nano 6, 6922–6929 (2012).
Paunovska, K. et al. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 18, 2148–2157 (2018).
Dahlman, J. E. et al. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl Acad. Sci. USA 114, 2060–2065 (2017).
Guimaraes, P. P. et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Release 316, 404–417 (2019).
Fenton, O. S. et al. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 29, 1606944 (2017).
Fenton, O. S. et al. Customizable lipid nanoparticle materials for the delivery of siRNAs and mRNAs. Angew. Chem. Int. Ed. 57, 13582–13586 (2018).
Sago, C. D. et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl Acad. Sci. USA 115, E9944–E9952 (2018).
Kauffman, K. J. et al. Rapid, single-cell analysis and discovery of vectored mRNA transfection in vivo with a loxP-flanked tdTomato reporter mouse. Mol. Ther. Nucl. Acids 10, 55–63 (2018).
Gilleron, J. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013).
Vermeulen, L. M. P. et al. Endosomal size and membrane leakiness influence proton sponge-based rupture of endosomal vesicles. ACS Nano 12, 2332–2345 (2018).
Demonte, D., Drake, E. J., Lim, K. H., Gulick, A. M. & Park, S. Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate. Proteins 81, 1621–1633 (2013).
Lim, K. H., Huang, H., Pralle, A. & Park, S. Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnol. Bioeng. 110, 57–67 (2013).
Kauffman, K. J. et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 15, 7300–7306 (2015).
Hassett, K. J. et al. Optimization of lipid nanoparticles for Intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids 15, 1–11 (2019).
Sabnis, S. et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
Hajj, K. A. et al. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small 15, e1805097 (2019).
Rui, Z. et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 9, 1449–1463 (2021).
Lokugamage, M. P., Sago, C. D., Gan, Z., Krupczak, B. R. & Dahlman, J. E. Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv. Mater. 31, 1902251 (2019).
Sago, C. D. et al. Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J. Am. Chem. Soc. 140, 17095–17105 (2018).
Miao, L. et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 11, 2424 (2020).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Chen, D. et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc. 134, 6948–6951 (2012).
Heyes, J., Palmer, L., Bremner, K. & MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 107, 276–287 (2005).
Acknowledgements
We acknowledge project funding from Translate Bio (Lexington, MA, USA) and the Marble Center for Cancer Nanomedicine, as well as support from the Cancer Center Support (core) (grant no. P30-CA14051) from the National Cancer Institute. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Biopolymers & Proteomics core and Preclinical Imaging and Testing core.
Author information
Authors and Affiliations
Contributions
L.H.R. developed the peptide barcoding concept. L.H.R., R.S.M. and D.G.A. conceived the study and designed experiments. L.H.R. and G.S. performed all in vitro and in vivo barcoding, hEPO and FLuc assays, including LNP formulation and animal work. A.K. developed and performed the LC–MS/MS method for all samples. R.S.M. designed and synthesized the combinatorial ionizable lipid library screened in this study. L.H.R., R.S.M. and D.G.A. contributed to the analysis and interpretation of the results and to the writing of the paper.
Corresponding author
Ethics declarations
Competing interests
Three of the authors (L.H.R., R.S.M. and D.G.A.) have filed a patent (US provisional application 63/289,343) on the technology described in this manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Xiangrong Song and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison of formulations containing DOPE or DSPC.
a Data of Fig. 4 replotted with DOPE formulations highlighted in red and DSPC formulations highlighted in blue. b Comparison of LNPs with identical molar compositions, differing only in phospholipid identity. Values shown are mean of n = 3 mice/group, with error bars representing the SD. * p < 0.05, ** p < 0.005, *** p < 0.0005.
Extended Data Fig. 2 Pilot screen using a small subset of the combinatorial library.
a A list of esters (left) and amines (right) used to generate lipids for the pilot screen. b Peptide barcoding analysis of the pilot library. Only lipids that resulted in a peptide dot product > 0.8 are shown. c Individual particle analysis using the top four hits, showing that the top three lipids from the peptide barcoding analysis do not result in significant protein production. d FLuc assay confirming that FLuc mRNA-containing LNPs composed of any of the three ‘false positives’ alone do not result in detectable protein expression, whereas a 1:1 mixture of hEPO mRNA-containing cKK-e12 LNPs and FLuc mRNA-containing ‘false positive’ LNPs results in robust protein expression. Values shown are the mean of n = 3 mice/group, with error bars representing the SD. The FLuc assay of panel d was performed twice with similar results.
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–5 and Tables 1–4.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rhym, L.H., Manan, R.S., Koller, A. et al. Peptide-encoding mRNA barcodes for the high-throughput in vivo screening of libraries of lipid nanoparticles for mRNA delivery. Nat. Biomed. Eng 7, 901–910 (2023). https://doi.org/10.1038/s41551-023-01030-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-023-01030-4
This article is cited by
-
Peptide codes for organ-selective mRNA delivery
Nature Materials (2026)
-
Multiplexed lipid nanoparticle barcoding reveals tissue-dynamic kinetic insights and enriched cellular tropism in hepatic zones
Nature Communications (2026)
-
Tissue-specific mRNA delivery and prime editing with peptide–ionizable lipid nanoparticles
Nature Materials (2026)
-
Using artificial intelligence to develop gene therapy for the lungs
Nature Biotechnology (2025)
-
Barcoded screening identifies nanocarriers for protein delivery to kidney
Nature Communications (2025)