Abstract
Messenger RNA vaccines have shown strong prophylactic efficacy against viral infections. Here we show that antigen-encoding small circular RNAs (circRNAs) loaded in lipid nanoparticles elicit potent and durable T cell responses for robust tumour immunotherapy after subcutaneous injection in mice, particularly when combined with immune checkpoint inhibition. The small circRNA vaccines are highly stable and show low levels of activation of protein kinase R as well as low cytotoxicity, enabling long-lasting antigen translation (longer than 1 week in cells). Relative to large protein-encoding unmodified or modified mRNAs and circRNAs, small circRNA vaccines elicited up to 10-fold antigen-specific T cells in mice and accounted for 30–75% of the total peripheral CD8+ T cells over 6 months. Small circRNA vaccines encoding tumour-associated antigens, neoantigens and oncoviral or viral antigens elicited substantial CD8+ and CD4+ T cell responses in young adult mice and in immunosenescent aged mice. Combined with immune checkpoint inhibition, monovalent and multivalent circRNA vaccines reduced tumour-induced immunosuppression and inhibited poorly immunogenic mouse tumours, including melanoma resistant to immune checkpoint blockade.
Similar content being viewed by others
Main
Conventional pathogen-based vaccines are associated with virulent reversion, genomic integration, limited biostability, anti-vector immunity, poor pharmacokinetics, weak immunogenicity, and limited benefits for cancer and chronic or pre-existing infections1,2. mRNA vaccines bypass some of these concerns2, as exemplified by the COVID-19 mRNA vaccines, which underscores their potential for versatile applications in disease prevention and therapy3,4. mRNA vaccines elicit innate immunity by the activation of pattern-recognition receptors (PRRs)5 and translate genuine antigens that elicit humoral responses or, upon proteolytic processing for optimal antigen presentation by major histocompatibility complex (MHC), elicit T cell responses. Nevertheless, there is still room to improve mRNA vaccines. For example, despite extensive structural and chemical modifications, current mRNA vaccines have (1) suboptimal biostability that limits the efficiency and duration of antigen translation and immunomodulation; (2) limited thermostability that causes short vaccine shelf-life and demands cold-chain storage and transportation; (3) heterogeneous mRNA impurities (such as double-stranded RNA (dsRNA), N – m, N + m), which are generated during manufacturing by in vitro transcription (IVT) and are difficult to completely remove during purification6; and (4) relatively long mRNA and mRNA impurities that may activate protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase and elicit overly strong innate immunity, and thus limit antigen translation efficiency, compromise prophylactic or therapeutic efficacies, and contribute to immunotoxicity7,8. These challenges also largely apply to self-amplifying mRNA that has long RNA replicons9. Furthermore, RNA replicons in self-amplifying mRNA can be sensitive to modifications and may interact with host factors or elicit anti-polymerase immunity that could neutralize future self-amplifying mRNA immunizations10. circRNA is an emerging class of RNA with unique biological functions11,12,13,14,15 and great potential for biomedical application. Natural circRNA is often generated from precursor mRNA back-splicing16. Recent attempts to develop synthetic circRNA therapeutics and vaccines17,18,19,20,21 rely on long mRNA associated with many of the above limitations.
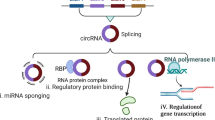
Here we report highly stable small circRNA vaccines that durably produce antigens, and thereby elicit robust and durable T cell responses with great safety for potent tumour combination immunotherapy (Fig. 1). Small circRNAs, typically less than 300 nucleotides, were designed to encode one or many peptide antigens and were synthesized by site-specifically ligating RNA oligonucleotides. An intrinsically fluorogenic aptamer-incorporated small circRNA was also synthesized to study circRNA stability. Small circRNA showed high thermostability (solution storage half-life of around 400 days at −20 °C) and biostability than not only the corresponding small linearized RNA but also state-of-the-art modified mRNA and large circRNA (typically larger than 1,000 nucleotides); furthermore, circRNA sequencing (circRNA-seq) verified the nucleotide authenticity of small circRNA transfected into live cells. The high stability of small circRNA enabled sustained peptide expression for at least a week in cells. Small circRNA vaccines activated the retinoic acid-inducible gene I (RIG-I) immunostimulation pathway in dendritic cells (DCs). Furthermore, compared with large nucleoside-modified mRNA and large circRNA, small circRNA vaccines showed good safety and low undesired PKR activation. Small circRNAs were efficiently loaded into nanocarriers, such as ionizable lipid nanoparticles (LNPs), for efficient delivery to antigen-presenting cells (APCs) in draining lymph nodes. Coupled with the innate immunostimulation by circRNA LNPs, the sustained antigen translation by small circRNA over a prolonged duration enables efficient and persistent antigen presentation and T cell priming. As a result, small circRNA vaccines encoding MHC-II- and MHC-I-restricted antigens elicited potent and durable (with memory) CD4+ and CD8+ T cell responses, respectively, both of which are pivotal for cancer therapy. Relative to multiple types of peptide/protein-encoding modified mRNA, unmodified mRNA and large circRNA, modification-free small circRNA vaccine elicited stronger and more long-lasting T cell responses that lasted at least 6 months post immunization. This makes small circRNA vaccines promising for in vivo generation of therapeutically significant doses of T cell receptor (TCR)-T cells, without any ex vivo T cell engineering and manufacturing. Moreover, small circRNA elicited profound T cell responses in immunosenescent aged mice. Modular small circRNA was easily adjusted to elicit T cell responses against various antigens, including tumour-associated antigens, tumour neoantigens, oncoviral antigens and viral antigens. In mice, small circRNA vaccines reduced tumour immunosuppression and enhanced tumour infiltration of antitumour immune cells. Monovalent or multivalent small circRNA vaccines, alone or combined with immune checkpoint blockade (ICB), notably inhibited or eradicated tumours in multiple syngeneic tumour models, including poorly immunogenic and ICB-resistant BrafV600E melanoma. These results demonstrate the great potential of small circRNA vaccines for tumour combination immunotherapy.
Small circRNA consists of peptide-antigen-encoding RNA and a short IRES and a Kozak consensus sequence that initiate peptide translation. Small circRNA was synthesized by ligating RNA oligonucleotide precursor(s) using RNA ligase and DNA splint(s). The absence of termini and the minimal sizes of small circRNA enable it to resist exonuclease degradation and minimize endonuclease degradation and hydrolysis, resulting in excellent thermostability and biostability. Nanocarriers delivered small circRNA vaccines to draining lymph nodes and intranodal APCs, in which (1) circRNA activates PRRs to elicit pro-inflammatory innate immunity with low PKR activation and low cytotoxicity, and (2) circRNA is efficiently translated to antigen peptides for antigen presentation over a prolonged duration. This allows small circRNA vaccines to elicit potent and long-lasting antigen-specific T cell responses. Modular small circRNA vaccines can be easily adjusted to encode various peptide antigens for versatile applications: (1) by encoding MHC-I- and MHC-II-restricted antigens, circRNA elicits CD8+ and CD4+ T cell responses, respectively; and (2) by encoding tumour-associated antigens, tumour neoantigens or (onco)viral antigens, circRNA holds the potential to develop off-the-shelf shared vaccines and personalized vaccines. Moreover, small circRNA vaccines reduce the immunosuppression and enhance the infiltration of antitumour immune cells in distant tumours. As a result, small circRNA vaccines, especially when combined with ICB, show robust immunotherapeutic efficacy for multiple types of tumour, including ICB-resistant BrafV600E melanoma.
Results
Optimization and characterization of highly stable small circRNA vaccines
Small circRNAs consist of minimal elements: codon-optimized peptide-antigen-encoding RNA and a short internal ribosome entry site (IRES) and a Kozak consensus sequence that recruit ribosomes for peptide translation (Fig. 1). Using MHC-I-restricted ovalbumin257–264 (OVA257–264 or SIINFEKL) as a model antigen, we prepared circRNA as follows: (1) circularizing 5′-phosphate-RNA using T4 RNA ligases and complementary 30-mer DNA splints; (2) removing linear/lariat RNA and DNA using DNase I and exonuclease T or RNase R22; (3) purification to remove immunostimulatory small DNA/RNA fragments, enzymes and nucleotides (Supplementary Fig. 1); and (4) verification by Sanger sequencing (Fig. 2a) and gel electrophoresis with an estimated circularization rate of 32% (Supplementary Fig. 1). A short IRES that efficiently recruits ribosomes and initiates translation is critical for the optimal immunomodulatory efficacy of small circRNA vaccines23. As shown by H-2Kb/SIINFEKL staining on vaccine-treated DCs, circRNA-SIINFEKL with 71-mer crucifer-infecting tobacco mosaic virus (crTMV) IRES enabled efficient antigen presentation, relative to circRNA-SIINFEKL with short IRES(es) long interspersed nuclear elements 1 (LINE1) and RNA binding motif protein 3 (RBM3), the corresponding linear RNA (liRNA, linearized circRNA with the nicking site between IRES and coding sequence, without 5′ cap or 3′ poly(A) or untranslated regions (UTRs) (Fig. 2a)), and CpG oligonucleotide-adjuvanted OVA protein24,25,26 (Fig. 2b,c, Supplementary Fig. 2a and Supplementary Table 1). Consistently, crTMV enabled circRNA-SIINFEKL to efficiently prime SIINFEKL-specific B3Z CD8+ T cell hybridoma (Fig. 2b–d). Deletion of IRES or Kozak sequence reduced antigen presentation and T cell priming (Supplementary Fig. 2b). Thus, crTMV IRES was selected for further studies. Interestingly, relative to state-of-the-art 5-methoxyuridine-modified OVA-encoding mRNA (5moU-mRNA-OVA), circRNA-SIINFEKL enhanced and prolonged antigen presentation and T cell activation (Fig. 2b,d and Supplementary Fig. 2b). A model Flag peptide-encoding circRNA (circRNA-Flag) efficiently expressed Flag products in live mouse DC2.4 cells, as shown by anti-Flag immunostaining (Fig. 2e).
a, Top: schematic illustration of the synthesis of small circRNA and liRNA by ligation of RNA oligonucleotide precursors and DNA splints. Bottom: Sanger sequencing of the cDNA of circRNA-SIINFEKL indicates precise and uniform ligation of RNA precursors into circRNA. The denoted RNA sequence is converted from the Sanger sequencing results of cDNA. b, crTMV IRES-based circRNA-SIINFEKL elicited efficient antigen presentation and T cell priming. Left: flow cytometric quantification of the mean fluorescence intensity (MFI) of SIINFEKL/H-2Kb complexes on DC2.4 cells treated with circRNA or controls for 24 h. Right: the activities of SIINFEKL-specific B3Z CD8+ T cell hybridoma co-incubated with the as-treated DC2.4 cells. B3Z cell activity: absorbance value. c, The secondary structures of crTMV IRES (left) and crTMV-based circRNA-SIINFEKL (right), as predicted using NUPACK. The blue box denotes IRES. d, Potent and durable priming of B3Z T cells by DC2.4 cells treated with circRNA-SIINFEKL. 5moU-mRNA-OVA: a benchmark protein-encoding 5moU-modified mRNA with CleanCap, 5′‑ and 3′‑UTRs, and 3′ A120. e, Confocal fluorescence microscopy images show the efficient production of a model peptide, Flag, from circRNA-Flag in live DC2.4 cells 24 h after transfection. f–h, circBroccoli was highly stable in live DC2.4 cells as shown by flow cytometry (f,g) and confocal fluorescence microscopy (h) analysis of cell Broccoli fluorescence intensities. Cells were transfected with circBroccoli or linear Broccoli control for 1–168 h, before adding fluorescence-activating Broccoli cognate, DFHBI-1T. Asterisks in g denote statistical significance of Broccoli MFI AUC of linear Broccoli relative to that of circBroccoli. i, circRNA-seq results showing the sequence integrity of circRNA-SIINFEKL recovered from live DCs after transfection for 24 h. circRNA-SIINFEKL in PBS was used as a positive control. The authenticity rate of each nucleotide was calculated as the rate of unmutated nucleotides. j, Percentages of intact circRNA-SIINFEKL, liRNA-SIINFEKL and 5moU-mRNA-OVA after storage in PBS at −20 °C, 4 °C or 23 °C for up to 180 days. At –20 °C, 4 °C and 23 °C, the half-lives of circRNA-SIINFEKL are estimated to be 401, 78 and 16 days, respectively; 143, 44 and 6 days, respectively, for 5moU-mRNA-OVA; and 2.6, <0.5 and <0.5 days, respectively, for liRNA-SIINFEKL. Data were quantified from gel electrophoresis using ImageJ. k, Upon transfection into DC2.4 cells, small circRNA-Flag showed durable Flag expression for at least 7 days, in contrast to fLuc expression for less than 3 days from 5moU-mRNA-fLuc. Intracellular Flag was stained using phycoerythrin (PE)-conjugated anti-Flag antibody, and the MFI of cells was measured by flow cytometry. l, Dynamic light scattering (left) and cryogenic electron microscopy (cryo-EM) images (right) showing the sizes and morphology of blank and circRNA-loaded ionizable SM-102 LNPs. m, Percentages of intact circBroccoli loaded in LNPs after storage in PBS at 4 °C or −20 °C (8% cryoprotectant sucrose) for 72 days. Data were quantified from flow cytometric analysis of DC2.4 cells transfected (24 h) with circBroccoli LNPs recovered from storage (paired t-test). n,o, Representative agarose gel electrophoresis images (n) and the intact circRNA percentages (o) of circRNA-SIINFEKL (1 nmol) and circRNA-RBD (1 nmol) after incubation in a series of diluted FBS in PBS (37 °C, 30 min). Data were quantified from gel electrophoresis by normalizing the band densities of FBS-treated circRNA to that of PBS-treated circRNA (t-test). RNA (100 nM) was transfected using Lipofectamine 3000 unless denoted otherwise. Data represent mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Bonferroni post-test.
mRNA can be degraded by ubiquitous biological and environmental RNases, especially exonucleases that are primarily responsible for mRNA degradation1, which limits mRNA shelf-life, biological half-life, antigen translation efficiency and duration, and consequently immunomodulatory efficacy. The absence of termini in circRNA prevents exonuclease degradation17. To study the biostability of circRNA in live cells, we synthesized a model circRNA in which the coding RNA was substituted with a fluorogenic RNA aptamer, Broccoli27. Upon transfection, the resulting circBroccoli showed excellent biostability in live DCs and the Broccoli fluorescence sustained for at least 7 days despite signal dilution during cell division, in contrast to almost complete fluorescence decay of linearized circBroccoli within 12 h (Fig. 2f–h and Supplementary Fig. 3a). Exogenous RNA, including circRNA, can be subject to mutation owing to RNA editing by endogenous editors, such as cytidine deaminases AID and APOBEC28, and adenosine deaminase ADAR29. Yet, such circRNA mutation may affect the ability of IRES and Kozak sequence to initiate peptide translation and alter peptide sequences. To study this, mouse bone marrow-derived dendritic cells (BMDCs) were transfected with circRNA-SIINFEKL. Twenty-four hours later, total circRNA was isolated from cells, and circRNA-seq analysis of circRNA-SIINFEKL showed no detectable circRNA mutations (Fig. 2i), suggesting the high sequence authenticity of this small circRNA in live cells. Modification-free small circRNA showed superior thermostability than not only the corresponding small liRNA but also 5moU-mRNA-OVA. Specifically, as estimated with a one-phase decay model, upon storage in phosphate-buffered saline (PBS) at –20 °C, 4 °C and 23 °C, the half-lives of circRNA-SIINFEKL are 401, 78 and 16 days, respectively, in contrast to 143, 44 and 6 days, respectively, for 5moU-mRNA-OVA, and 2.6, <0.5 and <0.5 days, respectively, for liRNA-SIINFEKL (Fig. 2j and Supplementary Fig. 3b). Consistently, when transfected into DCs, small circRNA-Flag showed durable Flag expression for at least 7 days when the experiment ended, in contrast to less than 3 days of protein expression by 5moU-mRNA-fLuc (Fig. 2k). Interestingly, circBroccoli-loaded LNPs (Fig. 2l) retained strong fluorescence intensity upon transfection in DCs after storage in solutions at 4 °C or −20 °C (supplemented with sucrose) for 70 days, verifying great thermostability of circRNA LNPs (Fig. 2m). circRNA is eventually degraded by hydrolysis30, endonuclease degradation31 and exonucleases upon circRNA linearization. Large circRNA has been often used for drug and vaccine development so far11. We hypothesize that because small circRNA typically has fewer sites of hydrolysis and endonuclease cleavage than large circRNA, small circRNA is overall more stable than large circRNA. To test this, we synthesized 1,760-mer large circRNA consisting of a long IRES (CVB3) and RNA encoding SARS-CoV-2 spike protein receptor-binding domain (RBD) (Supplementary Fig. 4). Upon incubation in diluted fetal bovine serum (FBS) followed by gel electrophoresis, 111-mer small circRNA-SIINFEKL showed higher stability than large circRNA-RBD (Fig. 2n,o). Overall, small circRNA showed excellent biostability and thermostability, which are critical for their long shelf-life, sustained antigen translation, and robust and durable immunomodulation.
LNPs efficiently delivered small circRNA to lymph nodes and APCs to elicit T cell responses
Nanocarriers are pivotal for the delivery and immunomodulation of mRNA vaccines32. To select a nanocarrier for small circRNA vaccines, we tested liposomes and three LNPs (with ionizable lipids SM-102, Dlin-MC3-DMA and Dlin-KC2-DMA, respectively), two classes of the most successful nucleic acid nanocarriers thus far32. We loaded circRNA-SIINFEKL and control 5moU-mRNA-OVA into these nanocarriers (N/P ratio 6). SM-102 LNPs showed comparable encapsulation efficiencies for mRNA (86.3%) and circRNA (86.9%). Owing to the small size of small circRNA, 185 copies of circRNA-SIINFEKL were calculated to be loaded per LNP with 170 nm in diameters, in contrast to 14 copies of 5moU-mRNA-OVA per LNP. Upon subcutaneous injection at the tail base of C57BL/6 mice, H-2Kb-SIINFEKL tetramer staining showed that SM-102 LNPs of 5 μg circRNA-SIINFEKL (10–15% typical doses of T cell mRNA vaccines33) elicited the most frequent SIINFEKL+CD8+ T cells among peripheral blood mononuclear cells (PBMCs) (Fig. 3a,b). Furthermore, as shown by Luminex, among all formulations, SM-102 LNPs of circRNA-SIINFEKL elicited the lowest levels of systemic chemokines associated with reactogenicity, a common side effect of mRNA vaccines (Fig. 3c and Supplementary Fig. 5). This indicates the good safety potential of small circRNA vaccines. Taken together, SM-102 LNP was selected as the nanocarriers for small circRNA vaccines. In vivo imaging system (IVIS) imaging showed that, for at least 8 days after subcutaneous injection in mice, SM-102 LNPs enhanced the accumulation of IR800-labelled circRNA (IR800-circRNA) in draining lymph nodes, which harbour various lymphocytes and orchestrate immunomodulation (Fig. 3d–f). This provides the basis for sustained antigen expression and T cell priming over a prolonged duration. Lymph nodes harbour various APC subsets that have differential vaccine uptake and immunostimulation efficacies. We studied circRNA uptake by intranodal APCs by flow cytometry 24 h after subcutaneous injection of Cy5-circRNA LNPs in mice. LNPs efficiently delivered circRNA to intranodal APC subsets that are pivotal to present peptide epitopes and elicit antigen-specific T cell responses, including CD8+ T cell-priming CD11c+CD11b−CD8+ classical DCs (cDCs), CD4+ T cell-priming CD11c+CD11b+CD4+CD103−CD205+ cDCs and CD11c+CD11b−CD8−CD4−CD103+ migratory cDCs (Fig. 3g). Moreover, within only 0.5 h after transfection in cultured DCs, LNPs enabled rapid endosome escape of circRNA to the cytosol to allow peptide translation (Fig. 3h,i and Supplementary Fig. 6). It is worth noting that liposomes also efficiently delivered small circRNA to draining lymph nodes and intranodal APCs in mice (Extended Data Figs. 1 and 2) and elicited SIINFEKL-specific CD8+ T cell responses more efficiently than 5moU-mRNA-OVA and CpG-adjuvanted OVA (Extended Data Fig. 3a–c). Meanwhile, presumably owing to vaccine-induced immunostimulation, liposomal circRNA-SIINFEKL upregulated expression of the immune checkpoint programmed death receptor 1 (PD-1) on T cells, especially SIINFEKL-specific CD8+ T cells (Extended Data Fig. 3d–f), providing an opportunity for ICB combination therapy with circRNA vaccines. Consistently, liposomal circRNA vaccine-immunized mice resisted challenge with EG7.OVA tumour cells, without significant weight loss, indicating the good safety potential of these vaccines (Extended Data Fig. 3g,h). Overall, nanocarriers such as LNPs efficiently delivered small circRNA vaccines to lymph nodes and pivotal intranodal APCs to elicit potent T cell responses with great safety potential.
a, Design of nanocarrier screening for small circRNA vaccines, using circRNA-SIINFEKL as a model and 5moU-mRNA-OVA as a control. RNA: 5 μg, subcutaneous (s.c.) injection at the tail base of C57BL/6 mice (n = 5) on days 0 and 14. b, Tetramer staining on day 21 showed that SM-102 LNPs of circRNA-SIINFEKL elicited the highest frequency of PBMC SIINFEKL+CD8+ T cells among all these RNA nanoformulations. c, Luminex results of normalized serum cytokine and chemokine levels 12 h after booster immunization (day 14). SM-102 LNP-circRNA-SIINFEKL induced relatively low reactogenicity-associated chemokines. Each cytokine or chemokine level (X) was respectively normalized as follows: \({X}_{\mathrm{normalized}}=\frac{X-{X}_{\min }}{{X}_{\max }-{X}_{\min }}\). d–f, Upon s.c. injection at the foot pad of BALB/c mice (n = 5), SM-102 LNPs efficiently delivered IR800-circRNA-SIINFEKL (0.5 nmol) to draining popliteal lymph nodes (dLNs) (circled in d), as shown by whole-body IVIS imaging (d), quantified IR800 fluorescence intensities of popliteal dLNs (e) and tissue fluorescence intensities quantified from ex vivo IVIS imaging (f). g, Flow cytometry results showing the Cy5-circRNA+ APC subsets among total CD45+ cells in draining lymph nodes 24 h after s.c. injection of free Cy5-circRNA or Cy5-circRNA LNPs, respectively. h, Confocal microscopy images showing efficient LNP delivery of Cy5-circRNA to DC2.4 cells and rapid endosome escape of Cy5-circRNA, the latter of which was indicated by the cytosolic Cy5-circRNA outside endolysosomes (circRNA, 100 nM; treatment, 0.5 h). Inset: one cell. i, As quantified from the above confocal microscopy images, the Cy5-circRNA fluorescence signal intensity ratios of outside/inside (O/I) endolysosome suggest the rapid endosome escape of LNP-circRNA in DC2.4 cells. Liposomal circRNA (lipo-circRNA) served as a control.
Small circRNA mediated immunomodulation with low PKR activation
Various types of RNA have been found to elicit innate immunity by activating endosomal PRRs such as Toll-like receptor 3 (TLR3), TLR7, TLR8 or cytosolic PRRs (such as RIG-I and melanoma differentiation-associated protein 5 (MDA5))34. Combined with the immunostimulatory ability of nanocarriers (for example, LNPs), mRNA vaccines elicit innate immunity that provides APCs with pro-inflammatory cytokines and co-stimulation signals for antigen presentation and T cell priming. To investigate the impact of small circRNA vaccines on immunomodulation, we conducted RNA-seq genome-wide transcriptomic analysis of mouse BMDCs LNP-transfected with circRNA-SIINFEKL and 5moU-mRNA-OVA, respectively, for 24 h, with PBS or blank LNPs as controls. About 560 genes were grouped by their involvement in inflammation, migration, antigen uptake and presentation, PRRs (such as RIG-I-like receptors (RLRs), TLRs and C-type lectin receptors (CLRs)), MHC-I and MHC-II, or miscellaneous immune-related genes. From the hierarchical clustering of these differentially expressed genes (Fig. 4a,b), we identified upregulated PRRs and activated immune-related pathways by LNP-transfected circRNA relative to blank LNPs. Overall, relative to blank LNPs, circRNA-SIINFEKL showed low upregulation of the above immune-related genes. For example, circRNA upregulated the transcriptions of Ddx58 (also known as Rigi) and Clec4, whereas 5moU-mRNA-OVA activated RLRs (Ifih1 (also known as Mda5), Oas1b, Eif2s1 and Eif2ak2 (Pkr)) and Tlr3 (Fig. 4c) that are characteristic of long dsRNA/ssRNA PRR activation. KEGG pathway analysis further confirmed that RNA sensor genes such as Rigi were activated by circRNA-SIINFEKL (Supplementary Figs. 7–10). The differential PRR activation profiles between circRNA-SIINFEKL and 5moU-mRNA-OVA may be due to structural differences such as the absence of long dsRNA in small circRNA. Furthermore, circRNA-SIINFEKL matured DCs, as indicated by enhanced gene activities of MHC-I (H2-K1) and co-stimulatory molecules (such as Cd40, Cd80 and Cd86) (Fig. 4c), which was supported by the elevated expression levels of co-stimulatory factors CD80 and CD86 (Supplementary Fig. 11). Moreover, circRNA-SIINFEKL triggered a multifaceted shift to inflammation in DCs, with upregulation of NLR (Nlrc4) and pro-inflammatory cytokines (Tnf and Il12rb1). circRNA-SIINFEKL also upregulated some immunosuppressive mediators, which presumably act as endogenous negative regulators of innate immune activation. Some of these genes encode therapeutic targets, for example, programmed death ligand 1 (PD-L1; encoded by Cd274), that hold potential for combination therapy with circRNA vaccines. To validate PRRs for circRNA, HEK-Lucia RIG-I, HEK-Blue hTLR3 and HEK-Blue hTLR7 reporter cells were respectively transfected with circRNA-SIINFEKL with controls, including large circRNA-OVA, 5moU-mRNA-OVA and blank transfection agent, as well as RNA immunostimulant polyinosinic-polycytidylic acid (poly(I:C)) as a positive control. circRNA-OVA comprises IRES CVB3 and a coding sequence (CDS) for OVA, and was synthesized by IVT and permuted intron–exon I (PIE-I)-mediated RNA circularization (Supplementary Table 2 and Supplementary Fig. 12a,b). As a result, circRNA-SIINFEKL activated RIG-I, but not TLR3 or TLR7 (Fig. 4d). RIG-I-dependent circRNA sensing was further validated in model RM1 cells with knockout of interferon-β (IFNβ) promoter stimulator I (IPS-1), an adaptor required by RIG-I-mediated type I IFN induction (Fig. 4e). These results provide the basis for future full understanding of the immunomodulation mechanism by small circRNA vaccines. Lastly, relative to large circRNA-RBD and 5moU-mRNA-OVA, small circRNA-SIINFEKL showed low PKR phosphorylation (pPKR) in cells, indicating low PKR activation (Fig. 4f,g and Supplementary Fig. 12c,d). This suggests that, relative to large circRNA and mRNA often with long dsRNA, the minimal long dsRNA in small circRNA minimizes pPKR inhibition of protein translation. Consistently, relative to large circRNA-RBD, small circRNA-SIINFEKL resulted in low cytotoxicity in HEK293T cells (Fig. 4h). Overall, these results reveal the immunomodulatory mechanism of small circRNA vaccines with low PKR activation and low cytotoxicity.
a–c, Gene transcriptome analysis results from BMDCs LNP-transfected with circRNA-SIINFEKL, 5moU-mRNA-OVA and PBS, respectively (24 h). The log2-transformed fold change (FC) represents log2(ratio of the mean expression induced by vaccine relative to PBS) (n = 3). a, Transcription heatmaps of genes involved in inflammation, migration, antigen processing and presentation, TLRs, RLRs, CLRs and miscellaneous immune-related genes. b, Volcano plot of differentially accessible peaks between blank LNPs and circRNA-SIINFEKL LNPs. c, log2(FC) of specific genes of interest related to the indicated pathways. d, PRR reporter cell activities upon treatment with circRNA-SIINFEKL (100 nM, 24 h) and controls. Poly(I:C) served as a positive control. e, RT–PCR results of Ifnb levels in wild-type (WT) RM1 and RM1-IPS1-KO cells treated with circRNA-SIINFEKL (100 nM, 48 h). f, Western blot analysis of PKR and pPKR in HEK293T cells after transfection with PBS, small circRNA-SIINFEKL, large circRNA-RBD and poly(I:C) positive control (0.5 μg per well), respectively, for 4 h. g, Western blot intensity ratio of pPKR to β-actin in treated HEK293T cells. Relative phosphorylation is indicated, calculated as the band intensity ratio (X) of pPKR to β-actin and then normalized to the relative phosphorylation induced by no RNA treatment group. \({X}_{\mathrm{normalized}}=\frac{\left({\mathrm{Intensity}}_{\mathrm{pPKR}}/{\mathrm{Intensity}}_{{\beta -\mathrm{actin}}}\right){\mathrm{for}}\; {\mathrm{RNA}}}{\left({\mathrm{Intensity}}_{\mathrm{pPKR}}/{\mathrm{Intensity}}_{{\beta -\mathrm{actin}}}\right){\mathrm{for}}\; {\mathrm{PBS}}}\). h, HEK293T cell viability transfected with the indicated RNA (0.5 μg per well, 24 h) or PBS control. Cell viability was normalized to PBS-treated cells. In d–h, RNA was transfected using Lipofectamine 3000. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by one-way ANOVA with Bonferroni post-test. NSm not significant.
Benchmarking small circRNA vaccines against modified mRNA, large circRNA and unmodified mRNA vaccines
Current mRNA vaccines use long poly(A) and artificial caps, as well as modified nucleotides for modified mRNA, which together result in balanced mRNA biostability, protein production and immunostimulation. Moreover, synthetic large circRNAs have been engineered as a novel class of mRNA vaccines against SARS-CoV-2 (ref. 17). We first studied the immunomodulatory efficacies of modification-free circRNA-SIINFEKL benchmarked against three forms of modified mRNA vaccines in vitro and in mice: (1) protein-encoding 5moU-mRNA-OVA, (2) SIINFEKL-encoding 5moU-mRNA (5moU-mRNA-SIINFEKL) and (3) pseudouridine (ψ)-mRNA-SIINFEKL. All these mRNAs have 5′/3′ UTRs, 3′ A120 and CleanCap (Supplementary Tables 1 and 2). After LNP transfection into DC2.4 cells, circRNA-SIINFEKL elicited the most rapid, most abundant and most durable SIINFEKL antigen presentation over 3 days (Fig. 5a and Supplementary Fig. 13). Next, C57BL/6 mice (6–8 weeks) were immunized with these RNAs at escalating doses (1, 3, 10, 30 and 100 μg), respectively, followed by monitoring their potency and duration of SIINFEKL-specific CD8+ T cell responses over 180 days, body weights and innate immune responses to evaluate T cell responses and vaccine safety (Fig. 5b). After two doses, tetramer staining (day 21) showed that while all these vaccines showed dose-dependent SIINFEKL+CD8+ T cell responses, circRNA-SIINFEKL elicited the largest fractions of SIINFEKL+CD8+ T cells among all PBMC CD8+ T cells (Fig. 5c and Supplementary Fig. 14). As low as 2 × 1 μg circRNA-SIINFEKL elicited 2.6- and 7.3-fold PBMC SIINFEKL+CD8+ T cell fractions relative to that by 5moU-mRNA-SIINFEKL and ψ-mRNA-SIINFEKL, respectively. Furthermore, at higher doses, circRNA-SIINFEKL caused less mouse body weight loss and more rapid weight recovery than benchmark mRNAs, indicating the potentially superior safety of small circRNA vaccines (Fig. 5d and Supplementary Fig. 15). A second booster of circRNA-SIINFEKL (day 28) substantially expanded the repertoire of SIINFEKL-specific CD8+ T cells, which plateaued at ~75% peripheral SIINFEKL-specific PBMC CD8+ T cells (day 35) at a dose of 3 × 30 μg (Supplementary Fig. 16). Over days 21–180, while all RNA vaccines showed dose-dependent SIINFEKL-specific T cell responses, the area under the curve (AUC) of SIINFEKL-specific PBMC CD8+ T cell fractions elicited by circRNA-SIINFEKL was higher than that by all the benchmark mRNA vaccines (Fig. 5e). Moreover, on day 35, the total counts of SIINFEKL+CD8+ T cells in peripheral blood and spleen per mouse induced by as low as 10 μg circRNA-SIINFEKL were estimated to be over 2-fold typical dose of adoptive TCR-T cells used for mouse solid tumour immunotherapy35 (Fig. 5f). This suggests the potential of small circRNA vaccines for in vivo generation of therapeutically significant dose of TCR-T cells for tumour immunotherapy, which bypasses any ex vivo TCR-T cell engineering and manufacturing. The T cell responses induced by circRNA-SIINFEKL waned less rapidly than that by modified mRNA, although high-dose 5moU-mRNA-OVA, which produces more immunogenic OVA than SIINFEKL, also elicited up to ~70% SIINFEKL-specific PBMC CD8+ T cells (day 35). Specifically, on day 180, 8–30% SIINFEKL-specific PBMC CD8+ T cells persisted with 10–100 μg circRNA-SIINFEKL, whereas such T cell fractions elicited by modified mRNA-SIINFEKL waned almost completely and that by 10-100 μg 5moU-mRNA-OVA waned to 3–20%. This suggests that circRNA-SIINFEKL elicited superior T cell memory than these benchmark mRNAs. The immune memory was verified by elevated total fractions of CD62L–CD44hi effector memory T (TEM) cells and D62L+CD44hi central memory T (TCM) cells, especially SIINFEKL+ memory T cells (Supplementary Fig. 17). Consistently, although the total counts of PBMC and splenic SIINFEKL+CD8+ T cells elicited by all vaccines reduced on day 180 relative to day 35, the T cell repertoires elicited by circRNA vaccines persisted for a longer duration than those elicited by benchmark mRNA (Fig. 5f). We expect that additional booster circRNA immunization would maintain the high T cell repertoires, when needed. Immunostimulation is often accompanied by immune exhaustion, as characterized by the upregulated expression of immune checkpoints such as PD-1. Levels of PD-1 in total PBMC CD8+ T cells in the immunized mice suggested dose-dependent PD-1 upregulation by day 35 (Supplementary Fig. 18), and PD-1 levels had largely recovered by day 49. The immune checkpoint upregulation can sensitize T cells for ICB, providing an opportunity to combine ICB with circRNA vaccines for optimal cancer therapeutic efficacy. Next, we benchmarked circRNA-SIINFEKL against unmodified OVA-encoding mRNA-OVA and large circRNA-OVA vaccines for their abilities to elicit T cell responses in mice. circRNA-OVA consists of an efficient long IRES (CVB3) and encodes highly immunogenic full-protein OVA (Supplementary Fig. 12a,b). Upon immunization in C57BL/6 mice (6–8 weeks), circRNA-SIINFEKL elicited higher fractions of PBMC SIINFEKL+CD8+ T cells than unmodified mRNA-OVA and large circRNA-OVA at equivalent doses over 120 days (Fig. 5g and Extended Data Fig. 4).
a–h, Benchmark studies of small circRNA vaccine versus three types of modified mRNA vaccine, an unmodified mRNA vaccine (no nucleoside modification), and a large circRNA vaccine in DCs and young adult mice. a, Three-day AUC of the H-2Kb-SIINFEKL MFI on DCs LNP-transfected with circRNA-SIINFEKL and three modified mRNAs, respectively. Data were quantified from flow cytometry results. Asterisks: statistical significance relative to circRNA. b, Timeline of in vivo benchmark study. C57BL/6 mice (6–8 weeks) were immunized with circRNA-SIINFEKL, as well as three modified mRNAs, unmodified mRNA-OVA and large circRNA-OVA, at escalating doses, respectively. c–e, Benchmarking circRNA-SIINFEKL against three types of modified mRNA for their ability to elicit T cell responses in mice (n = 9 for PBS, circRNA-SIINFEKL and 5moU-mRNA-OVA; n = 4–5 for the other groups). c, Tetramer staining results on day 21 suggest that circRNA-SIINFEKL elicited higher fractions of PBMC SIINFEKL-specific T cells than all modified mRNAs. circRNA-SIINFEKL elicited dose-dependent T cell responses, which plateaued at ca. 60% (dose 2 × 30 μg). Asterisks: statistical significance relative to circRNA. d, circRNA-SIINFEKL caused less mouse body weight drop and more rapid weight recovery than modified mRNAs, as exemplified 1–2 days after the 2nd 30 μg dose. e, 180-day kinetics of the PBMC SIINFEKL-specific CD8+ T cell percentages in the immunized mice, suggesting that circRNA-SIINFEKL elicited more potent and more durable T cell responses than these modified mRNAs, at all corresponding doses before the T cell responses plateaued. Asterisks denote statistical significance of the T cell fraction AUC relative to that for circRNA. f, Estimated total counts of peripheral and splenic SIINFEKL-specific CD8+ T cells per mouse (day 35, day 180). circRNA-SIINFEKL (3 × 10 μg) elicited SIINFEKL-specific CD8+ T cell counts that are >2-fold typical doses of adoptive TCR-T cells (5 × 106, dashed line) used for mouse tumour immunotherapy. g, Tetramer staining results on day 21 and day 35 suggest that circRNA-SIINFEKL elicited higher fractions of PBMC SIINFEKL-specific T cells than unmodified mRNA-OVA and large circRNA-OVA. Asterisks: statistical significance relative to circRNA. C57BL/6 mice (6–8 weeks) were subcutaneously immunized with circRNA-SIINFEKL (n = 9), unmodified mRNA-OVA (n = 4) and large circRNA-OVA (n = 4) (dose 3 μg and 10 μg, days 0, 14 and 28). h, Luminex heatmaps showing the serum chemokine and cytokine levels 6 h after the third immunization (day 28). Each cytokine or chemokine level (X) was independently normalized for each dose as follows: \({X}_{\mathrm{normalized}}=\frac{X-{X}_{\min }}{{X}_{\max }-{X}_{\min }}\). Overall, circRNA-SIINFEKL induced the least reactogenicity-associated chemokine and cytokine levels among all vaccines. i–k, Low-dose circRNA-SIINFEKL (5 μg; days 0, 14 and 28) elicited potent and durable T cell immunity in immunosenescent aged mice (1 year old; n = 5). i, Tetramer staining showed superior PBMC SIINFEKL-specific CD8+ T cell responses elicited by circRNA-SIINFEKL than 5moU-mRNA-OVA or CpG-adjuvanted OVA (day 21) (t-test). j, Intracellular IFNγ and TNF staining results in PBMC CD8+ T cells from the above immunized aged mice (day 35). T cells were restimulated with SIINFEKL peptide. k, circRNA-SIINFEKL vaccine protected immunized aged mice from EG7-OVA tumour cell challenge (1 × 106 cells, s.c. administration on day 70) (asterisks denote statistical significance relative to circRNA). Vaccines were delivered by SM-102 LNPs and subcutaneously injected at mouse tail base. Data represent mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by one-way ANOVA with Bonferroni post-test, unless denoted otherwise.
To study vaccine-elicited innate immunity associated with reactogenicity and pro-inflammation, we measured a panel of serum chemokines and pro-inflammatory cytokines by Luminex 12 h after the third dosing. Overall, relative to modified mRNAs and large circRNA-OVA, small circRNA-SIINFEKL induced lower levels of chemokines and cytokines that are often associated with immunotoxicity (Fig. 5h and Supplementary Fig. 19). This suggests the good safety potential of small circRNA vaccines. Overall, these results demonstrate the superior or at least comparable T cell responses and safety of small circRNA vaccines relative to modified mRNA and large circRNA vaccines.
Immunosenescence during ageing makes older people vulnerable to immune disorders and impairs vaccines’ immunomodulatory efficacy36. Thus, vaccines are highly desired to elicit potent and durable immunity in older people. In aged C57BL/6 mice (1 year old) (Fig. 5i,j), circRNA-SIINFEKL (5 μg, days 0 and 14) elicited 184% more frequent PBMC SIINFEKL+CD8+ T cells than 5moU-mRNA-OVA and 242% more than CpG-adjuvanted OVA (day 21), with potent T cell polyfunctionality and immune memory that protected mice from EG7.OVA cell challenge (Fig. 5i–k and Supplementary Fig. 20). These results suggest the ability of small circRNA vaccines to potentiate the T cell responses with memory in immunosenescent aged mice.
Broad application of circRNA vaccines for various peptide immunogens
Modular small circRNA vaccines can be easily adjusted to encode versatile peptide immunogens for broad applications. Beside MHC-I-restricted immunogens, MHC-II-restricted immunogens elicit CD4+ helper and effector T cell responses that are crucial for disease prophylaxis and immunotherapy. In C57BL/6 mice, circRNA encoding MHC-II-restricted OVA323–339 (named as ISQ) (5 μg; days 0 and 14) elicited ISQ-specific polyfunctional CD4+ T cells, as shown by intracellular IFNγ/TNF (formerly known as TNFα) staining (day 21) (Fig. 6a,b). Interestingly, circRNA-ISQ enhanced the ability of MHC-I-restricted circRNA-SIINFEKL to elicit SIINFEKL+CD8+ T cell response, likely because ISQ+CD4+ helper T cell response promoted SIINFEKL+CD8+ T cell priming37 (Fig. 6c). Moreover, circRNA-ISQ + circRNA-SIINFEKL outperformed 5moU-mRNA-OVA, which encodes OVA protein with both SIINFEKL and ISQ epitopes, to elicit PBMC SIINFEKL+CD8+ T cell response. Furthermore, small circRNA vaccines are applicable to tumour-associated antigens and tumour neoantigens for cancer immunotherapy. For example, circRNA encoding MHC-I-restricted ADPGK, an MC38 tumour cell-specific neoantigen38, elicited dose-dependent T cell responses with memory in mice (Fig. 6d,e and Supplementary Fig. 21). circRNA-ADPGK (2 × 5 μg) elicited 5-fold ADPGK +CD8+ T cells, relative to liRNA-ADPGK or CpG-adjuvanted ADPGK peptide; meanwhile, circRNA-ADPGK elevated the PD-1 level on CD8+ T cells, which can sensitize these cells for anti-PD-1 ICB in cancer combination immunotherapy (Fig. 6f). Multivalent vaccines can overcome the challenges of tumour antigenic heterogeneity and tumour cell immune evasion39. Bivalent circRNA encoding MHC-I-restricted melanoma-associated antigens mouse TRP2180–190 and human gp10023–33, the latter priming T cells to recognize both human and mouse gp100 (ref. 40), elicited bispecific CD8+ T cells (Fig. 6g). Lastly, T cell vaccines hold great potential to prevent and treat pathogenic viral infections and associated cancers, such as SARS-CoV-2 and human papillomavirus (HPV) infections, and HPV-associated cancers41. Relative to long B cell epitopes that are often susceptible to mutations, T cell peptide epitopes are short (8–20 amino acids) and highly conserved owing to less susceptibility to mutations. This makes T cell epitopes attractive to elicit broadly responsive T cell responses against rapidly mutating pathogens, such as SARS-CoV-2. As a step to this end, circRNA encoding MHC-I-restricted HPV16 E749–57 (ref. 41) outperformed liRNA-E749–57 and CpG-adjuvanted E749–57 peptides to elicit potent and long-lasting E749–57-specific CD8+ T cell response in mice (Fig. 6h,i and Supplementary Fig. 22). Likewise, circRNA encoding MHC-I-restricted SARS-CoV-2 spike protein RBD440–459 elicited potent and long-lasting RBD440–459+CD8+ T cells, as shown by tetramer staining over 70 days and intracellular cytokine staining (Fig. 6j,k). Overall, these results demonstrate the broad applicability of small circRNA vaccines to elicit T- cell responses.
a, Design of T cell response study for small circRNAs encoding various types of peptide antigen in C57BL/6 mice (6–8 weeks; n = 5). RNA, 5 μg per RNA; CpG, 5 μg; peptides, 10 μg; s.c. injection at tail base. Vaccines were delivered by LNPs. T cells were restimulated with OVA for intracellular cytokine staining. b, Intracellular IFNγ and TNF staining showed that MHC-II-restricted circRNA-ISQ elicited effector CD4+ T cells in PBMCs (day 21). 5moU-mRNA-OVA served as a benchmark. c, Intracellular IFNγ and TNF staining showed that MHC-II-restricted circRNA-ISQ enhanced the ability of MHC-I-restricted circRNA-SIINFEKL to elicit SIINFEKL-specific CD8+ T cell response (day 21). Both monovalent and bivalent circRNAs outperformed 5moU-mRNA-OVA to elicit the corresponding antigen-specific CD4+ or CD8+ T cell responses. d–k, Small circRNA vaccines elicited T cell responses against various cancer and viral peptide antigens. d, Tetramer staining results showed the fractions of ADPGK +CD8+ T cells among all live PBMC CD8+ T cells, indicating that circRNA-ADPGK neoantigen vaccine elicited robust T cell responses. e, circRNA-ADPGK elicited T cell memory (day 70). f, circRNA-ADPGK elevated PD-1 expression on CD8+ T cells (day 21). g, Intracellular IFNγ and TNF staining of PBMC CD8+ T cells (day 21) from C57BL/6 mice immunized with circRNA-TRP2/gp100 or CpG-adjuvanted peptide vaccines (days 0 and 14). h, The frequencies of PBMC E7+CD8+ T cells over 70 days post priming from C57BL/6 mice immunized with circRNA-E743–62 or CpG-adjuvanted peptide vaccines (days 0, 14 and 28). i, Intracellular IFNγ and TNF staining of PBMC CD8+ T cells (day 21) from the above immunized mice. j, Tetramer staining results showing the fractions of RBD440–459+CD8+ T cells among all live PBMC CD8+ T cells from as-immunized C57BL/6 mice (days 0, 14 and 28), indicating that circRNA-RBD440–459 elicited potent RBD440–459-specific CD8+ T cell responses. k, Intracellular IFNγ and TNF staining of PBMC CD8+ T cells (day 21) from the above immunized mice (asterisks in d, h and j: statistical significance relative to circRNA). Vaccines: subcutaneously injected at mouse tail base. Data represent mean ± s.e.m. (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by one-way ANOVA with Bonferroni post-test unless denoted otherwise.
Low-dose circRNA vaccines reduced tumour immunosuppression for potent immunotherapy
Tumour microenvironment (TME) is primarily where tumour cells suppress antitumour immunity2. In MC38 tumours, MC38-specific neoantigen vaccine circRNA-ADPGK, especially when combined with anti-PD-1, reduced immunosuppressive myeloid-derived suppressor cells (MDSCs) and CD4+FOXP3+CD25+ regulatory T (Treg) cells, increased the tumour infiltration of total CD8+ T cells and ADPGK+CD8+ T cells, and the ratio of enhanced CD8+ T cells to Treg cells in the tumour, which predicts tumour therapeutic efficacy (Fig. 7a–d). Consistently, RNA-seq of tumour tissue and transcriptome analysis revealed that circRNA-ADPGK and circRNA-ADPGK + anti-PD-1 triggered a multifaceted shift of the tumour to an inflamed and tumoricidal microenvironment, with upregulated gene transcripts related to pro-inflammatory cytokines, leukocyte-recruiting chemokines, factors associated with DC maturation and T cell priming, and natural killer (NK) cell and macrophage activation (Fig. 7e–g). These changes of the cellular and molecular tumour immune microenvironment are expected to be important attributes to the tumour therapeutic efficacy. As a result, circRNA-ADPGK neoantigen vaccine (3 × 5 μg) inhibited the growth of ADPGK-positive MC38 tumour (Fig. 7h). Moreover, circRNA-ADPGK potentiated the therapeutic efficacy of anti-PD-1, which blocks the upregulated PD-1 on energic T cells (Fig. 7h). Depletion of CD8+ T cells, but not CD4+ T cells or NK cells, abrogated the therapeutic efficacy of circRNA, verifying the central role of CD8+ T cells in MHC-I-restricted circRNA-ADPGK (Fig. 7i).
a, Study design of TME immune analysis and tumour immunotherapy in mice. MC38 tumour cells were subcutaneously inoculated in the flank of C57BL/6 mice, and treatment started when tumours reached around 60 mm3 on day 6. b–d, MC38 tumour immune microenvironment analysis (day 15) upon treatment with circRNA-ADPGK, alone or combined with anti-PD-1 (n = 6–8). 5moU-mRNA-ADPGK served as a control. b, The percentage of different immune cells among CD45+ cells in TME after the indicated treatment. c, Tetramer staining results showed the fractions of ADPGK+CD8+ T cells among total live CD8+ T cells in TME, indicating that circRNA-ADPGK vaccine combined with anti-PD-1 enhanced antigen-specific cytotoxic T cells within the TME. d, The ratio of CD8+ T cells to Treg cells within the TME. e–g, RNA-seq transcriptome analysis of MC38 tumours (day 15) after the above treatment (n = 3). e, Transcription heatmaps of selected genes related to immune modulation. f, The log-transformed mean expression ratio (log2(FC) of pathway-related genes in immunotherapy-treated tumours compared with that in PBS-treated tumours. g, Triwise radar plots depicting Gene Ontology enrichment analysis of T cell priming and DC activation pathways (left) and regulation of T cell proliferation (right). Black dots, all genes; red dots, genes related to the corresponding immune functions. h, Low-dose circRNA-ADPGK (5 μg) for potent combination immunotherapy of MC38 tumour, as shown by MC38 tumour growth and Kaplan–Meier mouse survival curves. liRNA and 5moU-mRNA encoding the same peptide antigen were used as controls (n = 14 for PBS, anti-PD-1, circRNA-ADPGK and circRNA-ADPGK + anti-PD-1; n = 7 for the other groups). Asterisks denote statistical significance between the AUC of circRNA-ADPGK tumour growth and that of circRNA-ADPGK + anti-PD-1. P = 0.0480: statistical analysis between the AUC of circRNA-ADPGK tumour growth and that of mRNA-ADPGK. i, MC38 tumour volumes after lymphocyte depletion using anti-CD8, anti-CD4 or anti-NK1.1 antibodies. RNA vaccines were delivered by SM-102 LNPs and subcutaneously injected at tail base. RNA, 5 μg; antibodies, 100 μg, intraperitoneal (i.p.) injection. Data represent mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by one-way ANOVA with Bonferroni post-test unless denoted otherwise.
Anticancer T cell responses and TME immunity are pivotal for effective cancer immunotherapy. Given their ability to elicit potent and durable T cell responses and reduce TME immunosuppression, small circRNA vaccines were further assessed for tumour immunotherapy in multiple mouse tumour models. Low-dose circRNA-SIINFEKL (3 × 5 μg) regressed EG7.OVA tumours (~50 mm3) and prolonged mouse survival, which outperformed benchmark 5moU-mRNA-OVA and CpG-adjuvanted OVA (Extended Data Fig. 4a–d). Depletion of CD8+ T cells, but not CD4+ T cells or NK cells, abrogated the therapeutic efficacy, verifying the central role of CD8+ T cells in MHC-I-restricted circRNA-SIINFEKL. circRNA did not cause changes in mouse body weights, indicating its good safety potential (Extended Data Fig. 4d). Moreover, small circRNA-SIINFEKL outcompeted full-protein OVA-encoding circRNA-OVA with a highly efficient IRES (CVB3) to inhibit EG7.OVA tumour growth in C57BL/6 mice (Extended Data Fig. 4e). High-risk HPV, including HPV 16, causes ~5% of all cancer cases. However, current US Food and Drug Administration (FDA)-approved HPV vaccines are unable to treat HPV-associated cancer. Moreover, current immunotherapy such as ICB and adoptive cell transfer are limited by ICB resistance and poor T cell memory. Therapeutic vaccines are promising to elicit T cell response for combination immunotherapy of HPV-associated cancer. MHC-I-restricted HPV16 protein E7 is an attractive oncoviral antigen because it is consistently and selectively expressed in HPV-associated cancer but not healthy cells, bypasses host immune tolerance, and is required to initiate and maintain HPV-associated cancer, which prevents cancer immune escape. Yet, current E7-based experimental therapeutic vaccines have limited efficacy against HPV-associated cancer in the clinic so far owing to low immunogenicity. Given the robust T cell response elicited by MHC-I-restricted circRNA-E7 (Fig. 6h,i), we tested circRNA-E7 for the immunotherapy of E7-positive TC-1 tumour. Low-dose circRNA-E7 (3 × 5 μg) inhibited TC-1 tumour progression via CD8+ T cells (Fig. 8b and Extended Data Fig. 4f). Because circRNA-E7 upregulated PD-1 on CD8+ T cells, combining circRNA-E7 with anti-PD-1 further enhanced the tumour therapeutic efficacy (Fig. 8b,c). We expect that increasing the circRNA dose would further improve the tumour therapeutic efficacy.
a, Design of tumour immunotherapy studies in mice. Tumour cells were inoculated subcutaneously in mouse flank, and treatment started when tumour volumes were around 50 mm3. Vaccines were loaded in LNPs and subcutaneously injected at tail base; antibodies were intraperitoneally injected. b, Average TC-1 tumour volumes after treatment with circRNA-E743–62 + anti-PD-1 and controls (n = 6–7). c, Spider plots of individual TC-1 tumour growth curves and complete regression (CR) rates after the above treatment. In b and c, vaccine, 5 μg RNA, 5 μg CpG, 10 μg E743–62 peptide antigens; anti-PD-1, 200 μg. Asterisks: statistical significance relative to circRNA + ICB. d, Average volumes of B16F10 melanoma treated with MHC-I/II-restricted tetravalent circRNA-T2/g/T1/T1 vaccine, alone or combined with anti-PD-1 + anti-CTLA-4. Asterisks denote statistical significance relative to circRNA-T2/g/T1/T1. e, B16F10 melanoma tumour growth after circRNA-T2/g/T1/T1 vaccine treatment and lymphocyte depletion. Asterisks: statistical significance relative to circRNA-T2/g/T1/T1. f, Spider plots of individual B16F10 melanoma tumour growth curves and CR rates. g, Kaplan–Meier survival curves of the as-treated B16F10 melanoma-bearing mice. Asterisks: statistical significance relative to circRNA + ICB. In d–g, circRNA, 10 μg; antibodies, 100 μg per ICB antibody, 200 μg anti-CD8, anti-CD4 or anti-NK1.1. h, Average (left) and individual (right) tumour growth curves of BrafV600E SM1 melanoma treated with circRNA-T2/g/T1/T1 combined with anti-PD-1 + anti-CTLA-4, as well as controls. Asterisks: statistical significance relative to circRNA-T2/g/T1/T1. i, Kaplan–Meier survival curves of BrafV600E SM1 melanoma-bearing mice treated as above. Asterisks: statistical significance relative to circRNA + ICB. In h and i, circRNA, 30 μg; ICB antibodies, 200 μg each. Vaccines: loaded in SM-102 LNPs and subcutaneously injected at mouse tail base. Data represent mean ± s.e.m. (n = 6–8). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by one-way ANOVA with Bonferroni post-test unless denoted otherwise.
Tumour antigenic heterogeneity and the resulting tumour cell immune escape represent a major obstacle against subunit vaccine-based tumour immunotherapy. Multivalent vaccines can at least partially overcome tumour antigenic heterogeneity and prevent tumour immune escape, thereby promoting tumour therapeutic efficacy. To test this using small circRNA vaccines, we engineered tetravalent small circRNA, termed as circRNA-T2/g/T1/T1, which encoded MHC-I-restricted mouse TRP2180–190, human gp10023–33 and mouse TRP1455–463, as well as MHC-II-restricted mouse TRP1106–130 (ref. 39). In poorly immunogenic B16F10 melanoma, MHC-I/II-restricted circRNA-T2/g/T1/T1 (3 × 10 μg) inhibited tumour progression, in which both CD8+ and CD4+ T cells were critical (Fig. 8d,e). Combining circRNA with both anti-PD-1 and anti-CTLA-4 further potentiated the therapeutic efficacy and prolonged mouse survival (Fig. 8d–g), with 2 out of 7 tumours showing complete regression without recurrence in 3 months (Fig. 8f). Around 50% of human melanoma contain BRAFV600E mutations, which cause constant oncogenic activation of BRAF, a serine/threonine protein kinase, and promotes melanoma tumorigenesis by, for example, evasion of antitumour immune responses, senescence and apoptosis. Syngeneic BrafV600E SM1 mouse melanoma is highly aggressive and poor immunogenic with highly immunosuppressive TME, making it often resistant to immunotherapy. Thus, SM1 melanoma closely resembles human BRAFV600E melanoma to predict its clinical therapeutic efficacy. Indeed, in syngeneic C57BL/6 mice, BrafV600E SM1 mouse melanoma did not respond to even dual ICB (anti-PD-1 + anti-CTLA-4). By contrast, tetravalent circRNA-T2/g/T1/T1 vaccine (3 × 30 μg) inhibited the progression of BrafV600E SM1 melanoma, and combining circRNA with anti-PD-1 + anti-CTLA-4 completely regressed 4 out of 8 tumours with 50% survival rate in 2 months (Fig. 8h,i). Overall, these results demonstrate the great potential of small circRNA vaccines for tumour combination immunotherapy.
Discussion
mRNA vaccines have been a great success as prophylactic COVID-19 vaccines and have shown promising clinical responses for cancer immunotherapy. Nonetheless, current mRNA vaccines are still associated with suboptimal stability, low loading capacity of very large mRNA in nanocarriers, heterogeneous by-products from IVT (such as truncated or extended mRNA, dsRNA), suboptimal immunomodulatory potency and duration, and reactogenicity. Addressing these issues may advance the development of mRNA vaccines, especially for the therapy of pre-existing diseases, such as cancer. To this end, we developed highly stable modification-free small circRNAs as mRNA vaccines to elicit potent and long-lasting T cell responses for robust tumour immunotherapy with great safety. Small circRNA is synthesized by ligating automatically synthesized RNA oligonucleotides, which could facilitate circRNA manufacturing and bypass purification complications associated with mRNA complex structures and mRNA by-products. Small circRNA has high loading capacity in nanocarriers for efficient delivery into draining lymph nodes and APCs. Small circRNA showed superior thermostability and biostability than not only small liRNA but also modified mRNA and large circRNA, the latter likely because of fewer sites of endonuclease cleavage and hydrolysis per small circRNA molecule than large circRNA. Moreover, circRNA-seq revealed the high nucleotide authenticity of circRNA in DCs, ruling out circRNA mutation under this condition owing to, for example, endogenous RNA editing. Highly stable small circRNA allowed for efficient and long-lasting antigen translation over at least 1 week. crTMV IRES enabled circRNA vaccine for efficient antigen presentation and T cell priming, presumably owing to strong ribosome binding with the structurally intact IRES in circRNA. The mechanism of peptide translation from small circRNA remains to be fully understood. Presumably, without a stop codon following peptide-coding RNA, circRNA is translated to a fused antigen and partial IRES-encoding peptide until reaching the intrinsic stop codons in crTMV IRES. Furthermore, if ribosomes skip discrete intrinsic stop codons42 in crTMV IRES, circRNA may produce long concatemeric antigenic peptides via rolling circle translation. Overall, these long peptide antigens would mimic ‘synthetic long peptides’ that promote adaptive immunity by authentic proteolytic processing and efficient antigen presentation.
Small circRNA elicited pro-inflammatory innate immunity and T cell responses. RNA-seq transcriptomic analysis verified that small circRNA activated a series of PRRs (for example, RIG-I), which provides the basis to further elucidate the innate immunomodulatory mechanisms for small circRNA vaccines. The ability and mechanisms of innate immunomodulation by small circRNA can be likely impacted by contexts, such as circRNA sequence and structure, nucleoside modifications, circRNA doses and pharmacokinetics, cell types and delivery systems. Compared with unmodified mRNA, which is typically highly immunostimulatory, unmodified small circRNA appears less immunostimulatory (comparably immunostimulatory relative to 5moU-modified mRNA) and more biostable, which is expected to permit long-lasting peptide expression by small circRNA. It is worth noting that circRNA and ionizable LNPs collectively mediate innate immunomodulation, which can promote antitumour and antiviral pro-inflammatory immunity but may also aggravate adverse side effects. Thus, it is desired to balance between the immunostimulation and the potential reactogenicity caused by circRNA LNPs for optimal clinical outcomes. In benchmarking studies, small circRNA vaccines outperformed several protein/peptide-encoding modified mRNA, unmodified mRNA and protein-encoding large circRNA to elicit potent and long-lasting antigen-specific T cell responses in mice. The corresponding PBMC and splenic antigen-specific T cell counts elicited by 3 × 10 μg small circRNA vaccines are estimated to be equivalent to the typical dose of adoptive TCR-T cells for mouse solid tumour immunotherapy. This provides an approach for in vivo production of therapeutically significant doses of antitumour TCR-T cells that can be boosted when needed, without any complex, costly and time-consuming ex vivo TCR-T cell engineering and manufacturing. Immune memory is pivotal for the long-term disease prophylactic and therapeutic response, as evidenced by ~1% chimeric antigen receptor (CAR) T cells in cured patients with cancer even years after CAR-T cell therapy43. Small circRNA vaccines elicited great T cell memory with large fractions of antigen-specific T cells that persisted for at least 6 months post immunization, which outperformed benchmark mRNA vaccines. Moreover, small circRNA elicited profound T cell response in immunosenescent aged mice. The ability of small circRNA vaccine to elicit potent and long-lasting T cell response was attributed to the high biostability of circRNA that prolonged its in vivo half-life and the synchronized innate immunostimulation and efficient antigen production over a prolonged duration. Lastly, compared with a modified mRNA and large circRNA, small circRNA showed low PKR activation, high cell viability and superior safety in mice.
Small circRNA vaccines can be easily adjusted to encode various types of peptide antigen for broad applications. Small circRNA encoding MHC-I-restricted or MHC-II-restricted antigens elicited CD8+ or CD4+ T cells, both pivotal for the prophylaxis and therapy of cancer and infectious diseases. Moreover, small circRNA vaccines elicited potent T cell responses against various disease-associated antigens such as tumour-associated antigens, tumour neoantigens, viral antigens and oncoviral antigens. Furthermore, small circRNA can be designed to express multiple antigens, which is critical for optimal tumour therapeutic efficacy by overcoming tumour antigenic heterogeneity and minimizing tumour immune evasion. The applicability of crTMV IRES-based circRNA in various antigens suggests the high structural integrity of this IRES in circRNA. We envision that engineering structurally rigid and strong ribosome-binding small IRES holds the potential to develop small circRNA for efficient peptide production in versatile applications. In mice, circRNA vaccines, especially when combined with ICB, remodelled the tumour immune microenvironment as indicated by the reduced frequencies of tumour immunosuppressive cells, enhanced tumour infiltration of antitumour immune cells and the overall transcriptomic shift to an antitumour pro-inflammatory status. Low-dose mono-/multivalent small circRNA vaccines exhibited robust tumour immunotherapy and enhanced the ICB therapeutic efficacy, including 50% of complete regression of ICB-resistant BrafV600E melanoma without recurrence in 2 months. Overall, small circRNA vaccines exhibited marked potential for tumour immunotherapy.
Compared with mRNA or large circRNA, small circRNA may hold advantages, including high stability, potent and long-lasting T cell responses, low PKR activation and good safety potential. Small circRNA vaccines may also find some limitations. For example, their application may be limited for large antigens that are often required to elicit humoral responses or for encoding many peptide antigens in the same circRNA vector to elicit multivalent T cell responses. Furthermore, owing to the limited length of RNA oligonucleotides synthesized by current automated RNA synthesizers, the synthesis of longer circRNA by ligating an increasing number of RNA oligonucleotides increases the complexity of circRNA manufacturing and may need to use alternative circRNA synthesis approaches (such as PIE systems44). Lastly, our results could spark future studies to further understand and develop small circRNA vaccines, including IRES–ribosome interaction in circRNA, peptide translation mechanisms, the immunological mechanisms underlying the robust and long-last immunity, and expanded applications such as infectious disease prevention and therapy.
Methods
circRNA synthesis
All oligonucleotides (sequences in Supplementary Table 1) were purchased from Integrated DNA Technologies. Small circRNAs were synthesized using T4 RNA ligase 1 or 2 (New England Biolabs) to ligate linear 5′-phosphorylated RNA precursors (5 μM each) in the presence of 30-mer DNA splints (15 μM each). The products were treated with DNase I and exonuclease T (New England Biolabs) or RNase R (Abcam) to remove DNA and linear or lariat RNA. circRNA-RBD and circRNA-OVA (sequence in Supplementary Table 1) were synthesized in two steps as reported previously44: (1) IVT using AmpliScribe T7 High Yield Transcription Kit (Biosearch Technologies, AS3107) and (2) group I intron-mediated autocatalytic circularization. In brief, 1 μg DNA template was used per 20 μl IVT reaction (37 °C, 2 h). Then, DNA templates were degraded with DNase I (37 °C, 15 min). The remaining RNA was column purified. Next, GTP (2 mM) was added to the resulting RNA to allow autocatalytic RNA cyclization (55 °C, 15 min). circRNA was column purified using an RNA Clean & Concentrator kit (Zymo Research) or HPLC to remove DNA or RNA fragments, nucleotides and enzymes. circRNA products were verified using gel electrophoresis, HPLC and Sanger sequencing of complementary DNA reverse transcribed from circRNA using primers flanking the circularization sites. Primers for circRNA-SIINFEKL reverse transcription: F, 5′-TTCGTTTGCTTTTTGTAGTATAATT-3′; R, 5′-AGTTTTTCAAAGTTGATTATACTCTCC-3′. Primers for circRNA-RBD reverse transcription: F, 5′-AAGCGGCTACATCCCAGAAG-3′; R, 5′-GGCGCACAAAGGTACCGTGA-3′.
In vitro circRNA stability
To study thermostability, small circRNA, liRNA and 5moU-mRNA-OVA (Trilink BioTechnologies, LLC) were stored in PBS at 4 °C, 25 °C and −20 °C for a series of times. Remaining RNAs were resolved by agarose gel electrophoresis and imaged using a ChemiDoc imaging system (Bio-Rad Laboratories). Intact RNA was quantified using ImageJ software (NIH). To study the in vitro biostability of large circRNA and small circRNA, RNA was incubated in freshly prepared diluted FBS (37 °C, 30 min) and was then briefly denatured in RNA loading buffer before gel electrophoresis. Intact RNA was quantified as above.
RNA LNP preparation
An ethanol phase containing all lipids and an aqueous phase of RNA were mixed to synthesize LNPs. The ethanol phase contained ionizable lipids, DSPC, DMG-PEG2000 and cholesterol at a molar ratio of 50:10:1.5:38.5. The aqueous phase contained circRNA or mRNA in 10 mM citrate buffer. The two phases were mixed at a flow rate of 1.8 ml min−1 and 0.6 ml min−1 (3:1), respectively, using Pump33DS syringe pumps. LNPs were dialysed in 1× PBS in a microdialysis cassette (20,000 molecular weight cut-off (MWCO), Thermo Fisher Scientific) at 4 °C for 12 h. LNP diameters and polydispersity index were measured on Zetasizer Nano (Malvern Instruments). RNA concentration and encapsulation efficiency in LNPs were measured using a modified Quant-iT RiboGreen (ThermoFisher) assay, and the circRNA copy numbers per LNP were estimated as before45. For cryo-EM, LNPs were concentrated after dialysis to ~90 mg ml−1 of lipid using Amicon ultracentrifugation filters before cryo-EM observation at the Molecular Electron Microscopy Core of University of Virginia.
circRNA LNP stability
Circular or linear Broccoli RNA aptamer loaded in LNPs was stored in PBS at 4 °C and −20 °C (supplemented with sucrose) for a series of durations. Then, recovered LNPs were transfected into DC2.4 cells for 1 h and were then resuspended and further incubated in PBS with 5% FBS and 200 μM DFHBI-1T (Sigma-Aldrich) for 45 min. Using untransfected cells as a control, the Broccoli fluorescence intensity (excitation, 488 nm; emission, 525 nm) of cells was analysed by flow cytometry.
Liposome encapsulation of circRNA
DOTAP, cholesterol and DSPE-PEG2000 (Avanti Polar Lipids) were mixed at molar ratios of 1:1:0.1 of DOTAP/cholesterol/DSPE-PEG2000. Lipid mixtures were dried under a nitrogen stream in glass test tubes. The resulting lipid films were placed in a desiccation system overnight. Dry films were hydrated in a solution of circRNA or mRNA in PBS with six cycles of vortexing for 30 s every 5 min. Samples were then freeze-thawed six times between liquid nitrogen and a 37 °C water bath and extruded using 21 passes in a mini-extruder (Avanti) through a 0.2 μm pore size polycarbonate filter (Whatman). Samples were placed in 10 K MWCO dialysis cartridges (Thermo Scientific) and dialysed against PBS for 6 h before use.
Cell culture
EG7.OVA cells were cultured in RPMI-1640 medium with 2 mM l-glutamine, 10% heat-inactivated FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 0.4 mg ml−1 G418. DC2.4, TC-1, SM-1 and B3Z cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 1× non-essential amino acids and 10 mM HEPES. DC2.4 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. B16F10, MC38 and HEK293T cells were cultured in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. All cells were cultured at 37 °C with 5% CO2.
BMDC isolation and culture
Mouse BMDCs were isolated from C57BL/6 mice (6–8 weeks). All femurs and tibia were collected and cut at the epiphysis level, and bone marrows were flushed out with 10 ml RPMI-1640 medium (Gibco) 3 times. The resulting cell suspensions were filtered through cell strainers (70 μm cut-off, BD Falcon), centrifuged and then lysed by ACK buffer (Gibco). To culture BMDCs, RPMI-1640 medium supplemented with 10% FBS and 20 ng ml−1 GM-CSF was added to resuspended cell pellets to obtain at a density of 2 × 106 viable cells per 75 mm Petri dish. Three days later, an additional 10 ml cell culture medium was added. Six days later, non-adherent and loosely adherent cells were collected by gentle washing with PBS and then pooled for further studies.
Cell viability assay
The cytotoxicity of DC2.4 cells (2 × 105 cells were seeded per well in 24-well plates) treated with small or large circRNAs was assessed using a ONE-Glo Tox kit (Promega, E7110). RNAs were transfected with Lipofectamine 3000 using serum-free medium 24 h after cell seeding. After 6 h, the medium was replaced by complete culture medium and the cells were incubated for another 18 h, before adding CellTiter-Fluor reagent. After incubation for 0.5 h, the fluorescence of cells was measured using a plate reader (Agilent BioTek) (excitation, 400 nm; emission, 505 nm).
Immunostaining of circRNA translation products in live DCs
Flag-encoding liRNA or circRNA (1 μg) was transfected into pre-seeded DC2.4 cells using Lipofectamine 3000 (Invitrogen). After 24 h, cells were washed and fixed using 4% paraformaldehyde in PBS at room temperature for 30 min and then incubated with 0.3% Triton X-100 in PBS. After blocking with 1% bovine serum albumin in PBS, cells were incubated with 2 μg ml−1 of anti-Flag M2 monoclonal antibody (Sigma-Aldrich) in PBS containing 1% bovine serum albumin for 1 h. Cells were washed twice with PBS containing 0.3% Triton X-100 and then incubated with a mixture of Alexa Fluor 488-labelled anti-mouse IgG antibody (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI; 250 ng ml−1) for 30 min. The cells were mounted on a glass slide using PermaFluor aqueous mounting medium (Thermo Fisher Scientific) for observation by confocal laser scanning microscopy on a Zeiss LSM 780 confocal microscope.
Intracellular translation kinetics of peptide and protein from mRNA and circRNA
DC2.4 cells were seeded into 24-well plates at a density of 2 × 105 cells per well and cultured at 37 °C for 24 h. Then cells were transfected with 5moU-mRNA-fFuc or circRNA-Flag (1 μg ml−1) using Lipofectamine 3000. For 5moU-mRNA-fFuc, after 24 h, 48 h and 72 h, the cell culture supernatants were replaced with a ONE-Glo Luciferase Assay System according to the manufacturer’s instructions, and the bioluminescence was measured using a microplate reader. For circRNA-Flag, the cells were collected, permeated and intracellularly stained with PE-conjugated anti-Flag antibody (BioLegend, 637309), followed by flow cytometric analysis of cell fluorescence intensity.
PKR activation
PKR activation was studied in mouse DC2.4 cells and human HEK293 cells. DC2.4 cells seeded in 24-well plates were treated with LNPs of circRNA-SIINFEKL or 5moU-mRNA-OVA (100 nM) for 24 h at 37 °C. Then, cells were collected and lysed with cell lysis buffer (Cell Signaling Technology) containing 100 mM phenylmethylsulfonyl fluoride (Thermo Fisher Scientific). Cell lysates were centrifuged at 12,000 rpm for 15 min at 4 °C and the supernatants were eluted with 1× SDS loading buffer and resolved by 10% SDS–PAGE, followed by western blotting using anti-PKR and anti-pPKR antibodies (Thermo Fisher Scientific). HEK293T cells were used to study PKR activation by small circRNA and large circRNA, with PBS and poly(I:C) as controls. Following RNA transfection, cells were lysed for 4 h, and proteins were resolved by SDS–PAGE, followed by western blotting to detect PKR and p-PKR using the above antibodies. Relative PKR phosphorylation was calculated as the gel density ratio of pPKR/β-actin, which was then normalized to the pPKR/β-actin gel density ratio in PBS-treated cells.
Flow cytometry
Flow cytometry was used to analyse immunostained cultured cells, mouse blood cells, as well as tumour tissue and lymph node homogenate single cells. Antibodies used for immunostaining are listed in Supplementary Table 3. Cultured cells were analysed on a BD Beckman Coulter flow cytometer. Blood and tissue single cells were analysed on LSRFortessa X-50 (BD Biosciences). Flow cytometry results were analysed using FlowJo V10 software. Representative flow cytometric analysis gating trees are shown in Supplementary Figs. 23–29.
Antigen presentation on cultured DCs
DC2.4 cells or BMDCs (2 × 105) seeded in 24-well plates were treated with circRNA-SIINFEKL or controls for 24 h. Cells were then collected and stained with APC-labelled anti-mouse H-2Kb/SIINFEKL complex antibody (BioLegend). Cells were then washed and analysed by flow cytometry.
B3Z cell activation
Upon recognition of the H-2Kb/SIINFEKL complex, B3Z cells (engineered SIINFEKL-specific CD8+ T cells) are activated and produce β-galactosidase. The activation level of the B3Z cells can be measured by the absorption of B3Z cell solution upon β-galactosidase treatment. Specifically, DCs were treated with SIINFEKL-specific vaccines for 16 h (RNA vaccine (1 μg) was transfected using Lipofectamine 3000). Cells were subsequently co-cultured with B3Z cells for another 24 h and were then lysed for 4 h at 37 °C with a lysis buffer (PBS with 100 mM 2-mercaptoethanol, 9 mM MgCl2, 0.2% Triton X-100 and 0.15 mM chlorophenol red-β-d-galactopyranoside). The reaction was stopped using a stop buffer (1 M sodium carbonate). B3Z T cell activation was quantified by measuring the absorbance at 570 nm with 635 nm as a reference wavelength. The B3Z cell activation is shown as the normalized optical density (OD) relative to the untreated cell control.
ELISA
DCs were plated at densities of 5 × 105 cells per well in 6-well plates. Cells were treated with the indicated formulations for 24 h. RNA (1 μg) was transfected into the cells using Lipofectamine 3000. Secreted cytokines (such as IFNβ, IL-6 and IL-12) in cell culture medium were quantified using ELISA kits (R&D Systems).
PRR activity studies using reporter cells
HEK-Lucia RIG-I cells, HEK-Blue hTLR3 cells or HEK-Blue hTLR7 cells were respectively seeded into a 96-well plate at a density of 3 × 104 cells per well and cultured at 37 °C for 24 h. Then cells were transfected with RNAs (1 μg ml−1) using Lipofectamine 3000. After 24 h, cell culture supernatants were collected and treated with either QUANTI-Blue Solution (InvivoGen, rep-qbs) or QUANTI-Luc 4 Lucia (InvivoGen, rep-qlc4lg1) according to the manufacturer’s instructions. The bioluminescence or OD at 630 nm was measured using a microplate reader.
In vitro cell uptake of circRNA
In vitro cell uptake of dye-labelled circRNA was examined using confocal laser scanning microscopy and measured by flow cytometry. Specifically, circRNA was labelled with Cy5 via hybridization with a Cy5-modified 30-mer cDNA. Cy5-circRNA loaded in LNPs or liposomes were incubated with DC2.4 cells for a series of durations and stained with LysoTracker Green DND-26 (Life Technologies) and 10 μg ml−1 Hoechst 33342 (Life Technologies) for 0.5 h. Cells were then washed with DPBS three times before confocal microscopy observation on a Zeiss LSM 780 confocal microscope. Alternatively, DC2.4 cells seeded in 24-well plates were treated with Cy5-labelled circRNA formulations as above, followed by flow cytometric analysis as above.
DC co-stimulation
Co-stimulatory factors CD80 and CD86 were stained and measured on vaccine-treated DC2.4 cells by flow cytometry. In brief, DC2.4 cells were treated with circRNA vaccine or controls, dissociated using non-enzymatic dissociation buffer, washed with PBS and resuspended in PBS with dye-labelled anti-mouse CD80 and CD86 antibodies at 4 °C for 30 min. Cells were then washed with PBS before flow cytometric analysis as above.
RNA-seq
Total RNA was isolated using AllPrep DNA/RNA Mini Kit (QIAGEN) with in-column DNase 1 digestion to obtain DNA-free RNA. The integrity of extracted RNA was analysed using the Agilent RNA 6000 Pico Kit (Agilent). An aliquot of 1 μg total RNA (RIN > 8) was used to prepare mRNA-seq libraries following the instruction of Illumina stranded mRNA-seq preparation Guide (Illumina). In brief, mRNA was captured by oligo (dT) magnetic beads from total RNA, cation fragmented and reverse transcribed into cDNA. cDNA was end repaired, ligated with RNA index anchor and PCR amplified to produce indexed libraries with IDT for illumined UD indexes. Following quality control, the resulting barcoded libraries were pooled in equal molarities and were paired-end (2 × 150 bp) sequenced on the NextSeq 2000 instrument (Illumina). The quality of RNA-seq reads was assessed with FastQC v0.11.9. Reads from individual samples were aligned using STAR aligner version 2.7.6a to mouse reference genome GRCm39. Raw gene counts of mapped reads were aggregated using featureCounts. The differential gene expression analysis was performed with Bioconductor package DESeq2 v1.30.0 using the normalized and filtered counts per gene from the RNA-seq data. Differentially expressed genes with adjusted P < 0.05 were subject to Gene Ontology and KEGG pathway enrichment analyses using the GSEA. The visualization of upregulated genes and associated Gene Ontology functions when comparing the three biological conditions was achieved using the Triwise R package. The Triwise R package enables two-dimensional visualization of differential gene expression patterns when comparing three biological conditions. It filters and calculates the average expression in the three biological conditions for each gene and then transforms this gene expression matrix to barycentric coordinates, reducing the three-dimensional matrix by one and retaining only the expression changes between samples. Limma package is then used to determine differentially expressed genes. These barycentric coordinates can then be plotted in a two-dimensional dot plot where the direction of a gene (a single dot) indicates in which condition(s) the gene is upregulated, while the distance from the origin represents the strength of upregulation.
circRNA-seq and circRNA variant calling for circRNA authenticity analysis
circRNA-seq was conducted for circRNA-SIINFEKL that was extracted from transfected cells using AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN), with untransfected circRNA-SIINFEKL as a control. circRNA libraries were constructed using 2 μg total RNA (RIN > 7), and ribosomal RNA was depleted using Ribo-Zero as well as an enzymatic depletion method (Illumina) and was purified with AMPure XP Beads (Beckman Coulter). Linear RNA was then digested using RNase R (Biosearch Technologies). The resulting circRNA was purified with AMPure XP Beads. circRNA was then cation fragmented at 95 °C for 2 min. Next, reverse transcription and library construction were performed using Illumina Stranded Total RNA Prep, followed by ligation with Ribo-Zero Plus Sample Preparation Guide (Illumina). Final libraries were size analysis by Agilent HS DNA chips (Agilent) and quantified by Qubit dsDNA HS Assay (Invitrogen). Following quality control, the resulting barcoded libraries were pooled in equal molarities and were paired-end (2 × 150 bp) sequenced on a NextSeq 2000 instrument (Illumina). The quality of raw FASTQ reads was assessed using FastQC v0.11.9. For read mapping, a reference fasta composed of two copies of circRNA-SIINFEKL sequence less one nucleotide was used. Reads were mapped either in a paired-end mode (PE) or in a single-end (SE) mode following read merging step. Here the reads were merged with either bbmerge function from BBTools package or merged reads (FLASh). The mapping step was performed with bowtie2 (bt2) with default settings, as well as with NextGenMap setting a constrain of minimum 90% identity (-i 0.9) over 75% of read length (-R 0.75) for a read to be considered as mapped. Finally, the variant calling step was done using the LoFreq tool. In brief, the mapping errors were corrected using ‘lofreq viterbi’, followed by indel quality insertion with ‘lofreq indelqual’, followed by base and indel alignment qualities insertions with ‘lofreq alnqual’. The final calling step was performed using ‘lofreq call’ with options set to ‘--no-default-filter --bonf 1 --sig 1 --call-indels’. To calculate the nucleotide authenticity rate of circRNA, reference fasta was divided into left and right flanks. Allele frequencies of left and right flank variants detected with LoFreq were summed and divided by the length of the flank. The resulting value was then used to calculate the nucleotide rates of circRNA.
Animal studies
All animal work was conducted in compliance to the Guide for the Care and Use of Animals under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Commonwealth University and University of Michigan. Vaccines were injected into mice subcutaneously either into mouse footpad for vaccine delivery study or into the mouse tail base for immune analysis or therapy studies. Both subcutaneous and intramuscular vaccine administrations expected to elicit comparable magnitude and quality of antigen-specific cellular or humoral immune responses46.
In vivo tissue and cell distribution of circRNA in mice
To study in vivo tissue distribution of circRNA, circRNA was labelled with IR800 via a 30-mer cDNA. IR800-circRNA loaded in LNPs (0.5 nmol circRNA) were subcutaneously administered in the foot pad of albino C57BL/6 mice (6–8 weeks). A series of days later, mice were imaged for IR800 fluorescence using an IVIS Lumina system (PerkinElmer). In another cohort, 24 h after administration, major organs were isolated and imaged on an IVIS Lumina system. The images were processed using Living Image analysis software (PerkinElmer).
To study intranodal cell distribution of circRNA by single cell flow cytometric analysis, C57BL/6 mice were subcutaneously administered with nanoparticles of 0.5 nmol Cy5-labelled circRNA at tail base. Twenty-four hours later, draining inguinal lymph nodes were collected, and lymph node single cells were prepared by passing minced lymph nodes through 70 μm cell strainers. Cells were then washed with and resuspended in cell staining buffer for staining using the following antibodies (BioLegend): Brilliant Violet 421 anti-mouse CD45, Alexa Fluor 594 anti-mouse CD11c, FITC anti-mouse CD11b, APC/Cy7 anti-mouse CD8a, PerCP/Cy5.5 anti-mouse CD4, Brilliant Violet 605 anti-mouse CD103, PE/Cy7 anti-mouse CD205, APC/Cy7 anti-mouse F4/80, PE/Cy5 anti-mouse CD11b and APC anti-mouse NK1.1. Zombie Aqua Fixable Viability Kit (BioLegend) was used to stain dead cells. Cells were then washed and analysed by flow cytometry.
Serum cytokine and chemokine measurement by Luminex
C57BL/6 mice were immunized, and blood was collected under isoflurane anaesthesia 12 or 24 h later. Blood was centrifuged for 5 min at 13,000 rpm, and sera were collected and stored at −80 °C. Pre-selected panels of cytokines and chemokines in the above samples were measured by Luminex (University of Virginia Flow Cytometry Core).
Tetramer staining on T cells
Mouse CD8+ and CD4+ T cells were stained for antigen-specific tetramers using PE-conjugated tetramers (NIH Tetramer Core Facility). In brief, peripheral blood was collected from the vaccinated mice and blood cells were enriched by centrifugation. Red blood cells were lysed using ACK lysis buffer for 10 min at room temperature. Blood clots were removed using a filter. Cells were washed twice in PBS and then stained using a Zombie Aqua Fixable Viability Kit (BioLegend) for 20 min at room temperature. Staining was quenched and cells were washed with FCS buffer (PBS buffer with 0.1% FBS). Cells were then blocked with anti-CD16/CD32 (BioLegend) for 10 min, followed by adding staining cocktail (PerCP/Cy5.5 anti-mouse CD3, APC/Cy7 anti-mouse CD8a, Brilliant Violet 421 anti-mouse PD-1 and tetramer-PE) and staining at room temperature for 30 min. Cells were then washed and 100 μl Cytofix was added into each well to resuspend cells before fixation at 4 °C for 20 min. Cells were then washed with Perm/Wash buffer and resuspended for flow cytometric analysis.
Intracellular cytokine staining in T cells
Peripheral blood was collected from immunized mice. Red blood cells were removed using ACK lysis buffer, and the obtained lymphocytes were transferred into U-bottom 96-well plates in 200 μl T cell culture media (RPMI-1640, 10% FBS, 100 U ml−1 penicillin/streptomycin, 50 μM β-mercaptoethanol, 1× MEM non-essential amino acid solution and 1 mM sodium pyruvate). Lymphocytes were pulsed with antigen peptides (40 μg ml−1) for 4 h, followed by addition of GolgiPlug Protein Transport Inhibitor containing brefeldin A (Thermo Fisher Scientific). The sequences of antigen peptides (CSBio) are shown in Supplementary Table 4. Cells were then placed in a culture incubator for 6 h before incubation with anti-CD16/CD32 for 10 min at room temperature. Cells were stained with APC/Cy7 anti-mouse CD8a, PerCP/Cy5.5 anti-mouse CD4 and Zombie Aqua Fixable Viability Kit for 20 min at room temperature. Cells were washed and subsequently fixed using Cytofix (BD Biosciences), washed, and permeabilized in 200 μl Cytoperm solution (BD Biosciences). Cells were then washed using Perm/Wash buffer (BD Biosciences), and permeabilized cells were then stained using PE anti-mouse IFNγ (BioLegend) and FITC anti-mouse TNF (BioLegend). Stained cells were washed for flow cytometric analysis.
Immune memory
Peripheral blood was collected from mice to analyse memory T cells. Immune memory was analysed by flow cytometric analysis of peripheral lymph node homing receptor, CD62L, and adhesion molecule, CD44. In brief, red blood cells were lysed using ACK lysis buffer, and blood cells were then collected by centrifugation and washing with FCS buffer. Cells were then blocked with anti-CD16/CD32 in FCS buffer for 10 min, followed by adding CD8-APC-Cy7, CD44-PE-Cy5, CD62L-FITC and dead cell-staining DAPI. Cells were stained at room temperature for 30 min and were then washed. Cytofix (100 μl) was added into each well to resuspend cells and cells were then incubated at 4 °C for 20 min. Cells were then washed with Perm/Wash buffer and resuspended for flow cytometric analysis. Central memory CD8+ T cells were analysed as CD44hiCD62Lhi CD8+ T cells; effector memory CD8+ T cells had variable to low levels of CD62L and high CD44 levels; and naive CD8+ T cells had high levels of CD62L and low to intermediate levels of CD44. To verify immune memory, immunized mice were challenged by s.c. inoculation of tumour cells (3 × 105) on the right shoulder. Tumour size was monitored every 3 days thereafter. Tumour volume was calculated using the formula volume = (length × width2)/2 and analysed using GraphPad Prism 7.
Tumour immunotherapy
About 3 × 105 EG7.OVA, MC38, TC-1, B16F10 and SM-1 BrafV600E cells, respectively, were subcutaneously inoculated on the shoulder of female C57BL/6 mice (6–8 weeks; The Jackson Laboratory; n = 6–8). Tumour growth was monitored by caliper measurement. Mice were euthanized when the maximal tumour dimension reached 2 cm, or the tumour volume exceeded 2,000 mm3, or developed ulceration. Mice were treated 6 days after tumour inoculation when tumours reached approximately 50 mm3. Typical doses: 5 μg RNA vaccine, 5 μg CpG + 10 μg antigen peptides or proteins, and 200 μg anti-PD-1 or anti-CTLA-4 (Bio X Cell, NH). Vaccine doses used in SM-1 BrafV600E melanoma therapy: 30 μg circRNA, 15 μg circRNA CpG and 30 μg antigenic peptides. Vaccines were subcutaneously injected at mouse tail base to allow lymphatic draining every 6 days three times, and anti-PD-1 or anti-CTLA-4 was injected intraperitoneally every 3 days five times. For lymphocyte depletion, anti-CD4, anti-CD8 and anti-NK1.1 (200 μg) were intraperitoneally injected every 3 days five times. Tumour size and mouse weight were monitored every 3 days. Tumour volumes were calculated and analysed as described above. Results were analysed using GraphPad Prism 7.
Statistical analysis
Data represent mean ± s.e.m., unless denoted otherwise. Statistical analysis was performed in GraphPad Prism Software version 5.0. P values were calculated by one-way ANOVA with Bonferroni post-test, unless denoted otherwise.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. Source data are available via figshare at https://doi.org/10.6084/m9.figshare.25919746. The raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request. Source data are provided with this paper.
Change history
12 August 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41551-025-01490-w
References
Rappuoli, R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 25, 1361–1366 (2007).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Pascolo, S. Vaccines against COVID-19: priority to mRNA-based formulations. Cells 10, 2716 (2021).
Teijaro, J. R. & Farber, D. L. COVID-19 vaccines: modes of immune activation and future challenges. Nat. Rev. Immunol. 21, 195–197 (2021).
Gholamalipour, Y., Karunanayake Mudiyanselage, A. & Martin, C. T. 3′ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses. Nucleic Acids Res. 46, 9253–9263 (2018).
Frederickson, R. & Herzog, R. W. RNA-based vaccines and innate immune activation: not too hot and not too cold. Mol. Ther. 29, 1365–1366 (2021).
Schwartz, S. L. & Conn, G. L. RNA regulation of the antiviral protein 2′-5′-oligoadenylate synthetase. Wiley Interdiscip. Rev. RNA 10, e1534 (2019).
Vogel, A. B. et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 26, 446–455 (2018).
Deering, R. P., Kommareddy, S., Ulmer, J. B., Brito, L. A. & Geall, A. J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 11, 885–899 (2014).
Liu, X. et al. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control. Release 348, 84–94 (2022).
Obi, P. & Chen, Y. G. The design and synthesis of circular RNAs. Methods 196, 85–103 (2021).
Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37, 667–675 (2019).
Liu, B. et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat. Biotechnol. 40, 1388–1393 (2022).
Yang, L., Wilusz, J. E. & Chen, L. L. Biogenesis and regulatory roles of circular RNAs. Annu. Rev. Cell Dev. Biol. 38, 263–289 (2022).
Chen, C.-y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417 (1995).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744 (2022).
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2022).
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
Tatomer, D. C. & Wilusz, J. E. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol. Cell 66, 1–2 (2017).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Fan, X., Yang, Y., Chen, C. & Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 13, 3751 (2022).
Chappell, S. A. & Mauro, V. P. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 278, 33793–33800 (2003).
Li, P. W., Li, J., Timmerman, S. L., Krushel, L. A. & Martin, S. L. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition. Nucleic Acids Res. 34, 853–864 (2006).
Skulachev, M. V. et al. Internal initiation of translation directed by the 59-untranslated region of the tobamovirus subgenomic RNA I2. Virology 263, 139–154 (1999).
Filonov, G. S., Moon, J. D., Svensen, N. & Jaffrey, S. R. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 136, 16299–16308 (2014).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
Li, Y. & Breaker, R. R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121, 5364–5372 (1999).
Thompson, J. E. et al. Limits to catalysis by ribonuclease A. Bioorg. Chem. 23, 471–481 (1995).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Kranz, L. M. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016).
Lamers, M. M., van den Hoogen, B. G. & Haagmans, B. L. ADAR1: “editor-in-chief” of cytoplasmic innate immunity. Front. Immunol. 10, 1763 (2019).
Arnaud, M. et al. Sensitive identification of neoantigens and cognate TCRs in human solid tumors. Nat. Biotechnol. 40, 656–660 (2022).
Yousefzadeh, M. J. et al. An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105 (2021).
Wang, J. C. & Livingstone, A. M. Cutting edge: CD4+ T cell help can be essential for primary CD8+ T-cell responses in vivo. J. Immunol. 171, 6339–6343 (2003).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Moynihan, K. D. et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat. Med. 22, 1402–1410 (2016).
van Stipdonk, M. J. et al. Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100. Cancer Res. 69, 7784–7792 (2009).
Nagarsheth, N. B. et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat. Med. 27, 419–425 (2021).
Mangkalaphiban, K. et al. Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae. PLoS Genet. 17, e1009538 (2021).
Fraietta, J. A. et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520 (2019).
Carrasco, M. J. et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 4, 956 (2021).
Ols, S. et al. Route of vaccine administration alters antigen trafficking but not innate or adaptive immunity. Cell Rep. 30, 3964–3971 (2020).
Acknowledgements
G.Z. acknowledges funding support from NIH (R01CA266981, R01AI168684, R35GM143014 and R21NS114455), DoD CDMRP Breast Cancer Breakthrough Award Level II (BC210931/P1), American Cancer Society Research Scholar Grant (RSG-22-055-01-IBCD), University of Michigan Rogel Cancer Center Discovery Award, NIH-NCATS KL2 scholarship (KL2TR002648) via Virginia Commonwealth University (VCU) C. Kenneth and Dianne Wright Center for Clinical and Translational Research (UL1TR002649), VCU Massey Cancer Center Molecules to Medicine pilot grant and VCU Commercialization Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Microscopy was performed at the VCU Microscopy Facility, supported in part by funding from NINDS Center Core Grant 5 P30 NS047463 and, in part, by funding from the NCI Cancer Center Support Grant P30 CA016059. Flow cytometry was performed at the VCU Massey Cancer Center Flow Cytometry Shared Resource, which is supported, in part, with funding from NCI Cancer Center Support Grant P30 CA016059. We thank the University of Michigan Flow Cytometry Core, the ULAM (Unit for Laboratory Animal Medicine) In Vivo Animal Core (IVAC), and the Advanced Genomics Core for service. We thank A. Ribas, J. Moon, C. Wu and NIH Tetramer Core for kindly providing cells and tetramer reagents.
Author information
Authors and Affiliations
Contributions
G.Z. conceptualized the project. G.Z. and Y.Z. designed the project. Y.Z., X.L., Q.W., Tinging Shen, S.Z., S.L., S.Y., Ting Su, L.M., L.X., Y.G. and C.G. performed the experiments. Y.Z., X.L., X.-Y.W. and G.Z. analysed the data. Y.Z., B.Z., K.H., K.M.T., X.Q., J.L. and G.Z. analysed the RNA-seq and circRNA-seq data. Y.Z., X.L. and G.Z. drafted and revised the paper, with assistance from K.H., S.Z., K.M.T. and J.L. All authors reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
G.Z. and Y.Z. are listed as inventors in a related patent application (WO2022173730A1). G.Z. is a co-founder, central scientific officer and equity holder of AmpedRNA Biosciences, LLC. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Christopher Jewell, Bowen Li and Jinjun Shi for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Liposomes promote circRNA delivery and antigen presentation in DCs.
a, DLS data showing the size distribution of blank liposome and lipo-circRNA with different N/P ratios in PBS (solid) and 1% FBS (dotted). b, Zeta potential of blank liposome and lipo-circRNA. c-d, In vitro cell uptake of circRNA and antigen presentation in DCs mediated by lipo-circRNA. c, Flow cytometry results and d, MFI of the SIINFEKL presentation in SIINFEKL-circRNA-treated DC2.4 cells (24 h). SIINFEKL antigen presentation on DC2.4 cells that were treated with Lipofectamine 3000-transfected circRNA and lipo-circRNA with different N:P ratios, respectively. Ns: non-significant, *p < 0.05, ***p < 0.001, by one-way ANOVA with Bonferroni post-test. e-h, In vitro intracellular delivery of circRNA in DCs by liposome. e, Confocal microscopy images of DC2.4 cells treated with lipo-circRNA for 1 h, 3 h, or 6 h. Blue: nuclei stained with Hoechst33342. Green: endolysosome stained with LysoTracker Green DND-26. Red: Cy5-circRNA. Insets: close-up views of single cells. f, Flow cytometry results of DC2.4 cells incubated with Lipo-circRNA and free circRNA for different times. g, Flow cytometry results showing MFI of DCs incubated with free or Lipo-Cy5-circRNA. ****p < 0.0001, by t-test of the AUC. h, The signal ratio of Cy5-circRNA outside/inside (O/I) the endolysosome.
Extended Data Fig. 2 Liposomes promoted the delivery of circRNA to draining lymph nodes and to key intranodal APC subsets in mice.
a, Liposomes promoted the delivery of IR800-circRNA to draining popliteal lymph nodes (circled) in BALB/c mice (0.2 nmol, s.c. injected at foot pad). b, AUC of the radiance efficiency. **p < 0.01, by t-test of the AUC. c, Ex vivo fluorescence images of BALB/c mice after s.c. injected at tail base for 24 h. d, Radiance efficiency of major organs and inguinal lymph nodes after s.c. injection of IR800-circRNA at tail base for 24 h. He: heart; Li: liver; Sp: spleen; Lu: lung; Ki: kidney; and LN: inguinal lymph node. e, The frequency of circRNA+ cDCs and macrophages in inguinal lymph node tissues 24 h after s.c. injection at tail base.
Extended Data Fig. 3 Small circRNA-SIINFEKL elicited potent T-cell responses in young adult mice.
a, Study design. C57BL/6 mice (n = 5) were vaccinated on day 0 and day 14, and PBMC T cell responses were analyzed starting from day 21. b, Tetramer staining on day21 – day70 showed that circRNA-SIINFEKL elicited potent SIINFEKL-specific CD8+ T cell response in mice (n = 5) that outperformed current benchmarks 5moU-modified CleanCap mRNA-OVA and CpG-adjuvanted OVA. c, circRNA-SIINFEKL elicited SIINFEKL-specific T cell memory (day 70). Tem: effector memory T cell; Tcm: central memory T cell. d, circRNA-SIINFEKL upregulated PD-1 expression on SIINFEKL-specific CD8+ T cells on day21. e, PD-1 MFI on live PBMC CD8+ T cells. f, circRNA-SIINFEKL enhanced PD-1 expression levels (MFI) and frequencies on SIINFEKL+CD8+ T cells than that on total CD8+ T cells in peripheral blood on day 21, indicating immune exhaustion often resulting from chronic immunostimulation and providing an opportunity to combine circRNA vaccines with immune checkpoint blockade for optimal T cell responses (Two-tailed paired t test). g, circRNA-SIINFEKL enabled mice to resist 3×105 EG7.OVA tumour challenge 70 days post-vaccination. Vaccine delivery by liposome, s.c. injected at tail base, 5 μg RNA, 2 nmole CpG, 20 μg OVA. *: relative to circRNA. h, Mouse body weights after tumour challenge. Data represent mean ± SD (b-e) and mean ± s.e.m. in other figure panels (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, by one-way ANOVA with Bonferroni post-test unless denoted otherwise.
Extended Data Fig. 4 Peptide-encoding small circRNA vaccine elicit stronger and more durable T-cell responses than protein-encoding unmodified mRNA and large circRNA vaccines in young adult mice.
a, Design of T cell response study in mice. C57BL/6 mice (n = 4-5; 6-8 weeks) were immunized with circRNA-SIINFEKL, unmodified mRNA-OVA, and large circRNA code OVA protein at 3 μg and 10 μg doses, respectively. b, 120-day kinetics of the PBMC SIINFEKL-specific CD8+ T cell percentages in the above immunized mice, suggesting that circRNA-SIINFEKL elicited overall stronger and more durable T cell responses than OVA-encoding unmodified mRNA and large circRNA vaccines. Asterisks: statistical significance of the T cell fraction AUC relative to that for circRNA. c, circRNA-SIINFEKL elicited larger fractions of memory T cells (Tem + Tcm) than unmodified mRNAs (day 90), indicating great T cell memory elicited by small circRNA vaccine. Data were quantified from CD44 and CD62L staining of PBMC CD8+ T cells.
Extended Data Fig. 5 Monovalent small circRNA vs modified mRNA or large circRNA vaccines for robust tumour immunotherapy.
a, For immunotherapy studies in mouse models of subcutaneous EG7.OVA (b-d), and TC-1 (e), tumours were inoculated into the right flank of C57Bl/6 mice. Vaccine: 5 μg RNA or 5 μg CpG + 10 μg protein or peptide antigens, s.c. injected in liposome at mouse tail base; antibodies: 200 μg, i.p. b-d, EG7.OVA tumour growth (b) and Kaplan-Meier survival curves (c) of EG7.OVA tumour-bearing mice treated with circRNA-SIINFEKL, 5moU-mRNA-OVA, and CpG-adjuvanted OVA, respectively. αCD8, αCD4, and αNK1.1 were injected intraperitoneally (i.p.) for lymphocyte depletion. d, Body weights of EG7.OVA tumour-bearing mice treated with circRNA-SIINFEKL vs. controls. e, Individual (upper panel) and average (lower panel) EG7.OVA tumor growth curves in C57BL/6 mice treated with the indicated circRNA-SIINFEKL or large circRNA-OVA, as well as αPD-1 (i.p.) alone or combined with circRNA-SIINFEKL. RNA: 30 μg, s.c. injection at tail base. CR: complete regression rate. f, TC-1 tumour volumes after lymphocyte depletion using αCD8, αCD4 or αNK1.1. Data represent mean ± s.e.m. *p < 0.05; **p < 0.01; ***p < 0.001, by one-way ANOVA with a Bonferroni post-test (n = 6-8).
Supplementary information
Supplementary Information
Supplementary methods, figures, tables and references.
Source data
Source Data Figs. 1–8 and Source Data Extended Data Figs. 1–5
Source data and statistics.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Liu, X., Shen, T. et al. Small circular RNAs as vaccines for cancer immunotherapy. Nat. Biomed. Eng 9, 249–267 (2025). https://doi.org/10.1038/s41551-025-01344-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-025-01344-5
This article is cited by
-
Circular RNA-based HPV16 therapeutic vaccine elicits potent and durable antitumor immunity
Journal of Experimental & Clinical Cancer Research (2026)
-
Circular RNAs in human biology: from splicing noise to master regulators
Molecular Biology Reports (2026)
-
CircRNAs: functions and emerging roles in cancer and immunotherapy
BMC Medicine (2025)
-
Functions and applications of enzymes in nucleic acid nanotechnology
Journal of Nanobiotechnology (2025)
-
Reprogramming neural-tumor crosstalk: emerging therapeutic dimensions and targeting strategies
Military Medical Research (2025)