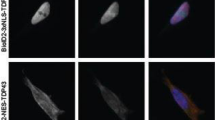
Abstract
In multiple neurodegenerative diseases, the RNA-binding protein TDP-43 forms cytoplasmic aggregates of distinct morphologies, including skein-like, small rounded granular and large spherical inclusions. Here, whereas the N-terminal self-oligomerization domain regulates TDP-43 demixing into cytoplasmic droplets, inhibition of N-terminal self-oligomerization domain-mediated oligomerization is shown to promote the formation of skein-like inclusions. Utilizing proximity labelling–mass spectrometry, cellular stresses are shown to induce TDP-43 association with actin-binding proteins that include filamins and α-actinin. Small interfering RNA-mediated reduction of filamin in Drosophila ameliorates cell loss from cytoplasmic TDP-43, consistent with the filamin–TDP-43 interaction enhancing cytotoxicity. TDP-43’s association with actin-binding proteins is mediated by BAG3, a HSP70 family nucleotide exchange factor that regulates the proteostasis of actin-binding proteins. BAG2, another HSP70 nucleotide exchange factor, facilitates the formation of small, rounded TDP-43 inclusions. We demonstrate that both TDP-43 self-oligomerization and its binding partners, including HSP70 and cochaperones BAG2 and BAG3, drive the formation of the different types of TDP-43 inclusion.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The quantitative mass spectrometry raw data have been deposited to the public database MassIVE with the dataset identifier MSV000098098 and ProteomeXchange with identifier PXD068255. The data can be downloaded from ftp://MSV000098098@massive-ftp.ucsd.edu. A previously published laser captured motor neuron RNA-seq dataset is available from the Gene Expression Omnibus under accession code GSE76220 (ref. 33). Previously published human spinal cord snRNA-seq datasets are available from the Gene Expression Omnibus under accession codes GSE190442 (ref. 44), GSE222322 (ref. 44) and GSE228778 (ref. 45) and at searchable website https://vmenon.shinyapps.io/humanspinalcord/ (ref. 45). Further information on the postmortem samples analysed are available from the UCSD ALS tissue repository (contact information please find in https://health.ucsd.edu/specialties/neuro/specialty-programs/als-clinic/pages/default.aspx). The images of in situ hybridization data for BAG3, BAG1, BAG2, BAG4, BAG5 and HSPH1 are from the Allen mouse spinal cord atlas46 (https://mousespinal.brain-map.org/imageseries/show.html?id=100020020, https://mousespinal.brain-map.org/imageseries/show.html?id=100024593, https://mousespinal.brain-map.org/imageseries/show.html?id=100038126, https://mousespinal.brain-map.org/imageseries/show.html?id=100037229, https://mousespinal.brain-map.org/imageseries/show.html?id=100048120, https://mousespinal.brain-map.org/imageseries/show.html?id=100021840). More representative images are available via Figshare at https://figshare.com/s/91c0eaea2f9c1f14a4b0 (ref. 75). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Ling, S.-C., Polymenidou, M. & Cleveland, D. W. J. N. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Nelson, P. T. et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 142, 1503–1527 (2019).
Josephs, K. A. et al. TAR DNA-binding protein 43 and pathological subtype of Alzheimer’s disease impact clinical features. Ann. Neurol. 78, 697–709 (2015).
McKee, A. C. et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J. Neuropathol. Exp. Neurol. 69, 918–929 (2010).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 63–70 (2006).
Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006).
Cohen, T. J. et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
McGurk, L. et al. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 (2018).
Wang, A. et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 (2018).
Gasset-Rosa, F. et al. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 (2019).
Mann, J. R. et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8 (2019).
Conicella, A. E. et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl Acad. Sci. USA 117, 5883–5894 (2020).
Wang, C. et al. Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid–liquid phase separation. Mol. Cell 79, 443–458.e7 (2020).
Arseni, D. et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 601, 139–143 (2022).
Arseni, D. et al. TDP-43 forms amyloid filaments with a distinct fold in type A FTLD-TDP. Nature 620, 898–903 (2023).
Jiang, Y. X. et al. Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43. Nature 605, 304–309 (2022).
Schweighauser, M. et al. Age-dependent formation of TMEM106B amyloid filaments in human brains. Nature 605, 310–314 (2022).
Yu, H. et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309 (2021).
Gruijs da Silva, L. A. et al. Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation. EMBO J. 41, e108443 (2022).
Wang, P., Wander, C. M., Yuan, C. X., Bereman, M. S. & Cohen, T. J. Acetylation-induced TDP-43 pathology is suppressed by an HSF1-dependent chaperone program. Nat. Commun. 8, 82 (2017).
Lu, S. et al. Heat-shock chaperone HSPB1 regulates cytoplasmic TDP-43 phase separation and liquid-to-gel transition. Nat. Cell Biol. 24, 1378–1393 (2022).
Jiang, L. L. et al. The N-terminal dimerization is required for TDP-43 splicing activity. Sci. Rep. 7, 6196 (2017).
Afroz, T. et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 (2017).
Oiwa, K. et al. Monomerization of TDP-43 is a key determinant for inducing TDP-43 pathology in amyotrophic lateral sclerosis. Sci. Adv. 9, eadf6895 (2023).
Lam, S. S. et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Yu, D. et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 12, 763–765 (2015).
May, J. A. et al. GPIIb-IIIa antagonists cause rapid disaggregation of platelets pre-treated with cytochalasin D. Evidence that the stability of platelet aggregates depends on normal cytoskeletal assembly. Platelets 9, 227–232 (1998).
Straight, A. F. et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743–1747 (2003).
Baas, P. W. & Black, M. M. Individual microtubules in the axon consist of domains that differ in both composition and stability. J. Cell Biol. 111, 495–509 (1990).
Krach, F. et al. Transcriptome-pathology correlation identifies interplay between TDP-43 and the expression of its kinase CK1E in sporadic ALS. Acta Neuropathol. 136, 405–423 (2018).
Melamed, Z. et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019).
Klim, J. R. et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019).
Ritson, G. P. et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 30, 7729–7739 (2010).
Behl, C. The Co-Chaperone BAG3 in Health and Disease. Trends in Pharmacol. Sci. 37, 672–688 (2016).
Rauch, J. N. et al. BAG3 is a modular, scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins. J. Mol. Biol. 429, 128–141 (2017).
Fuchs, M. et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem. J. 425, 245–255 (2010).
Reid Alderson, T. et al. A weakened interface in the P182L variant of HSP27 associated with severe Charcot–Marie–Tooth neuropathy causes aberrant binding to interacting proteins. EMBO J. 40, e103811 (2021).
Minoia, M. et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta. Autophagy 10, 1603–1621 (2014).
Wang, H. Q. et al. Induction of BAG2 protein during proteasome inhibitor-induced apoptosis in thyroid carcinoma cells. Br. J. Pharmacol. 155, 655–660 (2008).
Ulbricht, A. et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr. Biol. 23, 430–435 (2013).
Yadav, A. et al. A cellular taxonomy of the adult human spinal cord. Neuron 111, 328–344 e327 (2023).
Gautier, O. et al. Challenges of profiling motor neuron transcriptomes from human spinal cord. Neuron 111, 3739–3741 (2023).
Henry, A. M. & Hohmann, J. G. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mamm. Genome 23, 539–549 (2012).
Meister-Broekema, M. et al. Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks. Nat. Commun. 9, 5342 (2018).
Rauch, J. N. & Gestwicki, J. E. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414 (2014).
Carrettiero, D. C. et al. Stress routes clients to the proteasome via a BAG2 ubiquitin-independent degradation condensate. Nat. Commun. 13, 3074 (2022).
Carra, S., Seguin, S. J., Lambert, H. & Landry, J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 283, 1437–1444 (2008).
Hahn, O. et al. Atlas of the aging mouse brain reveals white matter as vulnerable foci. Cell 186, 4117–4133.e22 (2023).
Schmid, B. et al. Loss of ALS-associated TDP-43 in zebrafish causes muscle degeneration, vascular dysfunction, and reduced motor neuron axon outgrowth. Proc. Natl Acad. Sci. USA 110, 4986–4991 (2013).
Janssens, J. et al. Investigating the role of filamin C in Belgian patients with frontotemporal dementia linked to GRN deficiency in FTLD-TDP brains. Acta Neuropathol. Commun. 3, 68 (2015).
Tsujikawa, K. et al. Actin-binding protein filamin-A drives tau aggregation and contributes to progressive supranuclear palsy pathology. Sci. Adv. 8, eabm5029 (2022).
Schmid, E. T., Schinaman, J. M., Liu-Abramowicz, N., Williams, K. S. & Walker, D. W. Accumulation of F-actin drives brain aging and limits healthspan in Drosophila. Nat. Commun. 15, 9238 (2024).
Fulga, T. A. et al. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 9, 139–148 (2007).
Elliott, E., Tsvetkov, P. & Ginzburg, I. BAG-1 associates with Hsc70·Tau complex and regulates the proteasomal degradation of Tau protein. J. Biol. Chem. 282, 37276–37284 (2007).
Gamerdinger, M. et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28, 889–901 (2009).
Carrettiero, D. C., Hernandez, I., Neveu, P. & Kosik, K. S. The co-chaperone BAG2 sweeps PHF insoluble tau from the microtubule. J. Neurosci. 29, 2151–2161 (2009).
Carrettiero, D. C., Hernandez, I., Neveu, P., Papagiannakopoulos, T. & Kosik, K. S. The cochaperone BAG2 sweeps paired helical filament-insoluble tau from the microtubule. J. Neurosci. 29, 2151–2161 (2009).
Sheehan, P. W. et al. An astrocyte BMAL1-BAG3 axis protects against alpha-synuclein and tau pathology. Neuron 111, 2383–2398.e7 (2023).
Venturelli, E. et al. BAG1 is a protective factor for sporadic frontotemporal lobar degeneration but not for Alzheimer’s disease. J. Alzheimers Dis. 23, 701–707 (2011).
Hu, S. et al. Molecular chaperones and Parkinson’s disease. Neurobiology of Disease 160, 105527 (2021).
Serlidaki, D. et al. Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J. Biol. Chem. 295, 7301–7316 (2020).
de Fuenmayor-Fernández de la Hoz, C. P. et al. Distal hereditary motor neuronopathy as a new phenotype associated with variants in BAG3. J. Neurol. 271, 986–994 (2024).
Irobi, J. et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 36, 597–601 (2004).
Evgrafov, O. V. et al. Mutant small heat-shock protein 27 causes axonal Charcot–Marie–Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 36, 602–606 (2004).
Tang, M., Ji, C., Pallo, S., Rahman, I. & Johnson, G. V. W. Nrf2 mediates the expression of BAG3 and autophagy cargo adaptor proteins and tau clearance in an age-dependent manner. Neurobiol. Aging 63, 128–139 (2018).
Fernandopulle, M. S. et al. Transcription-factor mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 79, e51 (2018).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
He, L., Diedrich, J., Chu, Y. Y. & Yates, J. R. III Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal. Chem. 87, 11361–11367 (2015).
Xu, T. et al. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 (2015).
Tabb, D. L., McDonald, W. H. & Yates, J. R. III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Park, S. K. et al. Census 2: isobaric labeling data analysis. Bioinformatics 30, 2208–2209 (2014).
TDP-43 skein-like inclusions formed by BAG3/HSP70 guided co-aggregation with actin binding proteins attached to actin filaments. Figshare https://figshare.com/s/91c0eaea2f9c1f14a4b0?file=57302453 (2025).
Acknowledgements
We thank J. Santini at the UCSD Microscopy Core and P. Guo at the UCSD Nikon Imaging Center for assistance with imaging and image analysis. We are grateful for helpful discussions and experimental support from A. Goginashvili, C. Chen, M. S. Beccari, J. Lopez-Erauskin and P. Trivedi from the Cleveland lab. D.W.C. acknowledges support from the NIH (grant nos. R01NS27036 and R01-NS121604) and the Nomis Foundation, J.R.Y. acknowledges support from the NIH (grant no. P41 GM103533), J.R. acknowledges Target ALS Foundation and K.Z. is supported by the Guangdong Basic and Applied Basic Research Foundation (grant no. 2023B1515020109). We acknowledge the UCSD School of Medicine Microscopy Core Grant grant no. P30 NS047101.
Author information
Authors and Affiliations
Contributions
S.L. and D.W.C. conceived the project and planned the experiments. S.L., S.Z. and S.O. performed the experiments; S.O., T.O., O.A.A. and J.R. performed the patient postmortem tissue analysis; J.K.D. and J.R.Y. provided mass spec analysis support; S.V.S. helped to provide neuronal cultures; P.H. and K.Z. performed the fly experiment; and O.A.-G. helped on single nucleus RNA-seq data analysis. All authors interpreted data. S.L., S.Z., P.H., K.Z. and O.A.Z. prepared figures. S.L. and D.W.C. wrote the manuscript with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Philipp Kahle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Skein-like inclusion formation does not rely on the RNA binding capacity of TDP-43, but the RRM1 and low complexity domains are required.
(a) Representative images of cells expressing TDP-43∆NTD-Clover treated with NaAsO2 for 1 to 4 hours. Cells are stained with G3BP1 to show stress granules from more than three biological replicates. An arrow indicates a cell with TDP-43∆NTD-Clover in both stress granules and skein-like inclusions at 2 hours of arsenite treatment. (b-e) Representative live images of cells expressing TDP-4390-414-Clover from more than three biological replicates (b), TDP-4390-414-2KQ-Clover from more than three biological replicates (c), TDP-43102-414-2KQ-Clover from more than three biological replicates (d), TDP-43NLSm-S48D-Clover from more than three biological replicates (e) before and after 4 hours of 100 µM NaAsO2 treatment. Black-white inverted fluorescent images are shown. n = 4 biological replicates representing 149, 214, 146, 215 cells (TDP-43NLSm-Clover) and 37, 55, 58, 37 cells (TDP-43NLSm-S48D-Clover), respectively. Error bar: SD. (f) FRAP analysis of TDP-43 skein-like inclusions induced by MG132 from more than three biological replicates. High magnification images show no fluorescence recovery of photo bleaching points within 1 min. (g) Representative images of MG132-induced skein-like TDP-43 inclusions preserved after mild cell permeabilization. (h-i) Representative live images of cells expressing TDP-43174-414-Clover (h) and TDP-43∆NTD-∆320-343-Clover (i) before and after 4 hours of 100 µM NaAsO2 treatment from more than three biological replicates. Black-white inverted fluorescent images are shown. N = 4 biological replicates for TDP-43NLSm-S48D-Clover. (j) Representative images of cells expressing TDP-43∆NTD-V5 under conditions of no stress, NaAsO2, or MG132 treatment from more than three biological replicates. TDP-43∆NTD-V5 is indicated by V5 staining.
Extended Data Fig. 2 Concentration-dependent regulation of formation of skein-like and granule-like TDP-43 inclusions.
(a) Schematic of the experiment design to test the presence of different levels of cytoplasmic full-length TDP-43 alongside consistent expression levels of TDP-43∆NTD-Clover. The aim is to determine how the balance between full-length TDP-43 and the C-terminal fragment influences the formation of different types of TDP-43 de-mixing structures. (b) Representative live images of cells expressing similar levels of TDP-43∆NTD-Clover and either low or high levels of full-length RNA-binding-competent TDP-43NLSm-mRuby2 before and after 4 hours of NaAsO2 treatment from more than three biological replicates. (c) Representative live images of cells expressing similar levels of TDP-43∆NTD-Clover and either low or high levels of full-length RNA-binding-incompetent TDP-43NLSm/2KQ-mRuby2 before and after 4 hours of NaAsO2 treatment from more than three biological replicates.
Extended Data Fig. 3 TDP-43 skein-like inclusions are associated with actin filaments and actin binding proteins.
(a) Representative image showing no enrichment of mIFP on arsenite-induced TDP-43 inclusions in iPSC-derived motor neurons. (b-c) Representative images of skein-like TDP-43 inclusions colocalized with filamin A (FLNA, detected by immunostaining) in the non-permeabilized or permeabilized cells from more than three biological replicates. (d) Representative images of skein-like TDP-43 inclusions colocalized with actin filaments stained by phalloidin from more than three biological replicates. (e) Representative images of skein-like TDP-43 inclusions colocalized with actinin, detected by immunostaining from more than three biological replicates. (f) Representative images of cells withs TDP-43 skein-like inclusions stained with Click-iT RNA imaging kit from more than three biological replicates.
Extended Data Fig. 4 TDP-43, actin binding proteins, and BAG3 are enriched in the insoluble fractions after cell exposure to proteotoxic stress.
(a) Schematic diagram illustrating the biochemical analysis of soluble and insoluble fractions of cells expressing TDP-43∆NTD-Clover or Clover. (b) Analysis of the levels of TDP-43∆NTD-Clover, endogenous TDP-43, FLNA, β-Actin, Actinin, BAG3, HSPA1A, GAPDH in the soluble fractions of cells under no stress, 100 µM NaAsO2 or 10 µM MG132 treatment. Data are from three biological replicates. (c) Analysis of the levels of TDP-43∆NTD-Clover, endogenous TDP-43, FLNA, β-Actin, Actinin, BAG3, HSPA1A, GAPDH in the insoluble fractions of cells under no stress, 100 µM NaAsO2 or 10 µM MG132 treatment. Data are from three biological replicates. (d) Quantification of the levels of TDP-43∆NTD-Clover, endogenous TDP-43 and FLNA in both soluble and insoluble fractions. n = 3 independent replicates. Error bar: SD. (e) Specific interaction of TDP-43 with FLNA and actin under NaAsO2 or MG132 stress conditions detected by immunoprecipitation.
Extended Data Fig. 5 BAG3 is associated with skein-like structures of TDP-43.
(a) Representative images of motor neurons from ALS patients displaying different types of TDP-43 pathological structures. A dot-plot quantifies the percentage of motor neurons exhibiting different types of TDP-43 pathological structures. Number of motor neurons quantified are 400, 164, 114, 105, 90, 144, 70, 70, 80 from nine patients. Error bar: SD. (b) Schematic representation of the interaction domains of HSP70, BAG3 and HSPB8, HSPB1. (c-e) Expression levels of BAG1, BAG2, BAG3, BAG4, BAG5, BAG6 and HSPH1 in different cell types within the adult human spinal cord. Data are adapted from scRNA-seq datasets published by Yadav, et al., in Nature Neuroscience44 (c, d) and by Gautier, et al., in Neuron45 (e). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. (f) Expression levels of BAG1, BAG2, BAG4, BAG5 and HSPH1 in different cell types within the adult mouse spinal cord. Images are ISH data from the Allen mouse spinal cord atlas (https://mousespinal.brain-map.org/, https://mousespinal.brain-map.org/imageseries/show.html?id=100024593, https://mousespinal.brain-map.org/imageseries/show.html?id=100038126, https://mousespinal.brain-map.org/imageseries/show.html?id=100037229, https://mousespinal.brain-map.org/imageseries/show.html?id=100048120, https://mousespinal.brain-map.org/imageseries/show.html?id=100021840).
Extended Data Fig. 6 An elevated level of BAG3 increases the formation of TDP-43 skein-like structure and the sensitivity of TDP-43DNTD-Clover expressing cells to HSP70 inhibition in a HSP70-binding dependent manner.
(a) Representative images showing the localization of endogenous BAG3 in cells stained with BAG3 antibody from more than three biological replicates. (b) Western blot analysis of BAG3 in cells without the mRuby2-tagged BAG3 transgene or in cells stably expressing BAG3mRuby2 or BAG3R480A-mRuby2. (c-d) Western blot demonstrating the reduction of BAG3 (c) and HSPB1 (d) following siRNA treatment. (e) Decrease of HSPB8 following siRNA treatment detected by immunostaining and quantification of mean fluorescent intensity in cells. Volcano plots show the mean fluorescent intensities of HSPB8 and HSPB1 in cells treated with siHSPB8 or control siRNA. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. (f) Schematic illustrating the assessment of BAG3 association with TDP-43∆NTD-Clover on skein-like structures. (g-h) Representative live images of cells co-expressing TDP-43∆NTD-Clover with BAG3mRuby2 (g) or BAG3R480A-mRuby2 (h) upon 100 µM NaAsO2 treatment. (i) Measurement of HSP70 activity before, during, and after the removal of arsenite stress from more than three biological replicates. Error bar: SD. (j) The effect of expressing BAG3mRuby2 and BAG3 R480A-mRuby2 on cell resistance to HSP70 inhibition. Data are from N = 3 biological replicates. Number of quantified cells per replicate are 136, 168 and 202 (TDP-43∆NTD-Clover, no Dox); 140, 140, 156 (TDP-43∆NTD-Clover, Dox); 275, 286, 258 (TDP-43∆NTD-Clover and BAG3mRuby2, no Dox); 234, 178, 233 (TDP-43∆NTD-Clover and BAG3mRuby2, Dox); 198, 161, 210 (TDP-43∆NTD-Clover and BAG3 R480A-mRuby2, no Dox); 175, 166, 164 (TDP-43∆NTD-Clover and BAG3 R480A-mRuby2, Dox), respectively. Error bar: SD.
Extended Data Fig. 7 A mild decrease of HSP70 activity promotes the formation of TDP-43 skein-like inclusions induced by proteotoxic stress.
(a) Assessment of HSP70 chaperone activity under different levels of HSP70 inhibitor by examining the disruption of nuclear TDP-43 liquid annisosome from more than three biological replicates. (b) Quantification of cells exhibiting large aggregated TDP-43 granules under different levels of HSP70 inhibitor treatment. N = 3 biological replicates. Cell numbers are 82, 117, 127 (5 μM VER155008); 114, 90, 144 (10 μM VER155008); 110, 86, 78 (50 μM VER155008). Error bar: SD. (c) Representative live images of cells expressing TDP-43∆NTD-Clover treated with 10 µM MG132 and low concentrations of HSP70 inhibitor. (d) Representative live images of cells expressing TDP-43∆NTD-Clover treated with 100 µM NaAsO2 and low concentrations of HSP70 inhibitor from more than three biological replicates. (e) Quantification of cells with skein-like TDP-43 inclusions under the conditions described in (c) and (d). N = 3 biological replicates. Cells number quantified per replicate are 205, 177, 220 (2 hr, NaAsO2, no HSP70i); 197, 159, 204 (3 hr, NaAsO2, no HSP70i); 210, 163, 211 (4 hr, NaAsO2, no HSP70i); 200, 225, 205 (2 hr, NaAsO2, 5 μM HSP70i); 225, 225, 224 (3 hr, NaAsO2, 5 μM HSP70i); 232, 217, 220 (4 hr, NaAsO2, 5 μM HSP70i); 234, 213, 172 (2 hr, NaAsO2, 10 μM HSP70i); 231, 211, 190 (3 hr, NaAsO2, 10 μM HSP70i); 253, 232, 197 (4 hr, NaAsO2, 10 μM HSP70i); 195, 207, 185 (2 hr, MG132, no HSP70i); 149, 138, 130 (3 hr, MG132, no HSP70i); 65, 86, 76 (4 hr, MG132, no HSP70i); 152, 173, 168 (2 hr, MG132, 5 μM HSP70i); 137, 138, 119 (3 hr, MG132, 5 μM HSP70i); 106, 96, 77 (4 hr, MG132, 5 μM HSP70i); 199, 203, 171 (2 hr, MG132, 10 μM HSP70i); 175, 113, 129 (3 hr, MG132, 10 μM HSP70i); 108, 74, 86 (4 hr, MG132, 10 μM HSP70i). Error bar: SD.
Extended Data Fig. 8 Association of BAG2 and BAG1L with TDP-43 de-mixed structures requires their ability to bind HSP70.
(a-c) Quantification of cells co-expressing TDP-43∆NTD-Clover and HSPH1mRuby2 (a), BAG1SmRuby2 (b) or BAG1MmRuby2 (c) that form skein-like inclusions, granule-like inclusions, or both types of inclusions after 4 hour of 100 µM NaAsO2 or 10 µM MG132 treatment. Number of cells quantified from one representative biological replicate: 284 cells (HSPH1mRuby2, NaAsO2), 247 cells (BAG1SmRuby2, NaAsO2), 252 cells (BAG1MmRuby2, NaAsO2); 255 cells (HSPH1mRuby2, MG132), 236 cells (BAG1SmRuby2, MG132), 193 cells (BAG1MmRuby2, MG132). (d) Representative live images of cells co-expressing TDP-43∆NTD-Clover and BAG1LmRuby2 before and after treatment with 10 µM MG132 and 100 µM NaAsO2 from more than three biological replicates. (e) Representative live images of cells co-expressing TDP-43∆NTD-Clover and BAG1L2RA-mRuby2 before and after treatment with 10 µM MG132 and 100 µM NaAsO2 from more than three biological replicates. (f) Quantification of cells from one representative biological replicate co-expressing TDP-43∆NTD-Clover and BAG1LmRuby2 that form skein-like inclusions, granule-like inclusions, or both types of inclusions after 4 hour of 100 µM NaAsO2 or 10 µM MG132 treatment. Number of cells quantified from one representative biological replicate: 230 cells in NaAsO2 group, 160 cells in MG132 group. (g) Representative live images of cells co-expressing TDP-43∆NTD-Clover and BAG2I60A-mRuby2 before and after treatment with 10 µM MG132 and 100 µM NaAsO2 from more than three biological replicates. (h) Representative live images of cells co-expressing TDP-43∆NTD-Clover and BAG2∆20-61-mRuby2 before and after treatment with 10 µM MG132 and 100 µM NaAsO2 from more than three biological replicates.
Supplementary information
Supplementary Tables 1–8
Supplementary Table 1. Significantly altered proteins identified in the proximity labelling dataset (Padj <0.01 and log2(fold change) >1 or <−1 in either the arsenite or MG132 treatment group). Supplementary Table 2. Identification results of proximity labelling of TDP-43∆NTD-Clover-APEX2 (whole list, no P value and fold change cut off). Supplementary Table 3. RNA levels of the candidates identified to associate with TDP-43∆NTD-Clover-APEX2 on skein-like inclusions in the laser-captured motor neuron transcriptome dataset. Supplementary Table 4. Plasmid information. Supplementary Table 5. Patient information (age). Supplementary Table 6. Patient information (disease course). Supplementary Table 7. Patient information (gender). Supplementary Table 8. Patient information (site of onset).
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig.4 2
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, S., Zhang, S., Oung, S. et al. TDP-43 skein-like inclusions are formed by BAG3- and HSP70-guided co-aggregation with actin-binding proteins. Nat Cell Biol 27, 1925–1937 (2025). https://doi.org/10.1038/s41556-025-01789-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01789-5