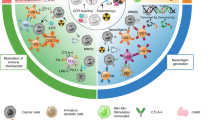
Abstract
Chemoresistance and immunosuppression are common obstacles to the efficacy of chemo-immunotherapy in colorectal cancer (CRC) and are regulated by mitochondrial chaperone proteins. Here we show that the disruption of the tumour necrosis factor receptor-associated protein 1 (TRAP1) gene, which encodes a mitochondrial chaperone in tumour cells, causes the translocation of cyclophilin D in tumour cells. This process results in the continuous opening of the mitochondrial permeability transition pore, which enhances chemotherapy-induced cell necrosis and promotes immune responses. On the basis of this discovery we developed an oral CRISPR–Cas9 delivery system based on zwitterionic and polysaccharide polymer-coated nanocomplexes that disrupts the TRAP1 gene in CRC. This system penetrates the intestinal mucus layer and undergoes epithelial transcytosis, accumulating in CRC tissues. It enhances chemotherapeutic efficacy by overcoming chemoresistance and activating the tumour immune microenvironment in orthotopic, chemoresistant and spontaneous CRC models, with remarkable synergistic antitumour effects. This oral CRISPR–Cas9 delivery system represents a promising therapeutic strategy for the clinical management of CRC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets of tumour transcriptome sequencing have been deposited in the The National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) with the accession number PRJNA1096614. Source data are provided with this paper.
References
Ward, R. A. et al. Challenges and opportunities in cancer drug resistance. Chem. Rev. 121, 3297–3351 (2021).
Galluzzi, L., Buqué, A., Kepp, O., Zitvogel, L. & Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 17, 97–111 (2017).
Thibaudin, M. et al. First-line durvalumab and tremelimumab with chemotherapy in RAS-mutated metastatic colorectal cancer: a phase 1b/2 trial. Nat. Med. 29, 2087–2098 (2023).
Zhou, C. et al. Outcomes and toxicities of immune checkpoint inhibitors in colorectal cancer: a real-world retrospective analysis. Cancer Commun. 41, 921–924 (2021).
Schmitt, M. et al. Colon tumour cell death causes mTOR dependence by paracrine P2X4 stimulation. Nature 612, 347–353 (2022).
Mariya, T. et al. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol. Res. 2, 1220–1229 (2014).
Giddings, E. L. et al. Mitochondrial ATP fuels ABC transporter-mediated drug efflux in cancer chemoresistance. Nat. Commun. 12, 2804 (2021).
Wu, Z. et al. Mitochondrial DNA stress signalling protects the nuclear genome. Nat. Metab. 1, 1209–1218 (2019).
Lao, L. et al. CD8+ T cell-dependent remodeling of the tumor microenvironment overcomes chemoresistance. Cancer Immunol. Res. 11, 320–338 (2023).
Kang, B. H. et al. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 131, 257–270 (2007).
Wang, X. et al. Cyclophilin D deficiency attenuates mitochondrial perturbation and ameliorates hepatic steatosis. Hepatology 68, 62–77 (2018).
Zhao, Q. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93.e22 (2020).
Denorme, F. et al. Platelet necrosis mediates ischemic stroke outcome in mice. Blood 135, 429–440 (2020).
Feng, Y., Madungwe, N. B., Tombo, N., Li, L. & Bopassa, J. C. Abstract 420: RIP3 interacts with mitofilin in the inner membrane of mitochondria to induce cardiomyocytes necrosis after ischemia/reperfusion. Circ. Res. 123, A420 (2018).
Costantino, E. et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 279, 39–46 (2009).
Yoon, N. G. et al. Mitoquinone inactivates mitochondrial chaperone TRAP1 by blocking the client binding site. J. Am. Chem. Soc. 143, 19684–19696 (2021).
Gewirth, D. T. Paralog specific Hsp90 inhibitors—a brief history and a bright future. Curr. Top. Med. Chem. 16, 2779–2791 (2016).
Tong, S., Moyo, B., Lee, C. M., Leong, K. & Bao, G. Engineered materials for in vivo delivery of genome-editing machinery. Nat. Rev. Mater. 4, 726–737 (2019).
Abramson, A. et al. Oral mRNA delivery using capsule-mediated gastrointestinal tissue injections. Matter 5, 975–987 (2022).
Yoo, J., Nhean, S., Vogel, P., Rybkin, I. I. & Kostoff, D. Implementation of oral chemotherapy management program in the large integrated health care system and its impact on patient safety. J. Clin. Oncol. 36, 279 (2018).
Brown, T. D., Whitehead, K. A. & Mitragotri, S. Materials for oral delivery of proteins and peptides. Nat. Rev. Mater. 5, 127–148 (2020).
Li, B. et al. Trimethylamine N-oxide-derived zwitterionic polymers: a new class of ultralow fouling bioinspired materials. Sci. Adv. 5, eaaw9562 (2019).
Shao, Q. & Jiang, S. Molecular understanding and design of zwitterionic materials. Adv. Mater. 27, 15–26 (2015).
Bansil, R. & Turner, B. S. The biology of mucus: composition, synthesis and organization. Adv. Drug Deliv. Rev. 124, 3–15 (2018).
Duncan, G. A., Jung, J., Hanes, J. & Suk, J. S. The mucus barrier to inhaled gene therapy. Mol. Ther. 24, 2043–2053 (2016).
Mirji, G. et al. The microbiome-derived metabolite TMAO drives immune activation and boosts responses to immune checkpoint blockade in pancreatic cancer. Sci. Immunol. 7, eabn0704 (2022).
Teft, W. A. et al. Identification and characterization of trimethylamine-N-oxide uptake and efflux transporters. Mol. Pharm. 14, 310–318 (2017).
Kato, Y. et al. Synthetic zwitterions as efficient non-permeable cryoprotectants. Comm. Chem. 4, 151 (2021).
Ganguly, P., Boserman, P., van der Vegt, N. F. A. & Shea, J.-E. Trimethylamine N-oxide counteracts urea denaturation by inhibiting protein–urea preferential interaction. J. Am. Chem. Soc. 140, 483–492 (2018).
Xie, W. J. et al. Large hydrogen-bond mismatch between TMAO and urea promotes their hydrophobic association. Chem 4, 2615–2627 (2018).
Landriscina, M. et al. TRAP1, a novel antiapoptotic gene responsible for multidrug resistance in human colorectal carcinoma cells. J. Clin. Oncol. 27, 2541 (2009).
Chae, Y. C. et al. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell 22, 331–344 (2012).
Ariës, I. M. et al. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J. Exp. Med. 215, 3094–3114 (2018).
Lu, L. et al. Nanoparticle-based oral delivery systems for colon targeting: principles and design strategies. Sci. Bull. 61, 670–681 (2016).
Hone Lopez, S. et al. The gut wall’s potential as a partner for precision oncology in immune checkpoint treatment. Cancer Treat. Rev. 107, 102406 (2022).
Lee, Y., Kamada, N. & Moon, J. J. Oral nanomedicine for modulating immunity, intestinal barrier functions, and gut microbiome. Adv. Drug Deliv. Rev. 179, 114021 (2021).
Zhao, Z. et al. Organoids. Nat. Rev. Methods Prim. 2, 94 (2022).
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018).
Li, X., Bechara, R., Zhao, J., McGeachy, M. J. & Gaffen, S. L. IL-17 receptor–based signaling and implications for disease. Nat. Immunol. 20, 1594–1602 (2019).
Overall, C. M. & Kleifeld, O. Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6, 227–239 (2006).
Zhao, H. et al. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol. Cancer 21, 144 (2022).
Pan, K. & Xie, Y. LncRNA FOXC2-AS1 enhances FOXC2 mRNA stability to promote colorectal cancer progression via activation of Ca2+-FAK signal pathway. Cell Death Dis. 11, 434 (2020).
Baghdadi, M. et al. Chemotherapy-induced IL34 enhances immunosuppression by tumor-associated macrophages and mediates survival of chemoresistant lung cancer cells. Cancer Res. 76, 6030–6042 (2016).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 52333004 to X.-Z.Z. and 22135005 to X.-Z.Z.).
Author information
Authors and Affiliations
Contributions
K.Z. and X.-Z.Z. conceived the project and designed the experiments. K.Z. synthesized materials. K.Z. and Y.Y. performed in vitro cell experiments. K.Z., Y.Y. and X.-K.J. contributed to data collection and analysis. K.Z., Y.Y., X.K.J., T.P., S.-M.Z., C.-H.Y. and Z.-Y.R. performed in vivo experiments. K.Z., Y.Y. and X.-Z.Z. cowrote the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Rene Jackstadt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Enhanced tumour uptake and penetration of HTPBD.
a, Representative CLSM 3D visualization images of CT26 cells in the lower chamber. Scale bar, 50 μm. b, Representative CLSM images showing colocalization of nanocomplexes and mucus in mouse colon after 15 min incubation. Scale bar, 200 μm. c,d, Representative CLSM images of nanocomplexes penetration into tumour cell clusters from apex to equator with 20 μm z-spacing (c); fluorescence intensity distribution of nanocomplexes at 60 μm depth (d). Scale bar, 50 μm.
Extended Data Fig. 2 HTPBDTRAP1 + 5-FU treatment significantly induced transcriptomic changes in genes associated with the immune system.
a, Venn analysis of CRC tissue transcription in control, HTPBDTRAP1, and HTPBDTRAP1 + 5-FU treatment groups. b,c, Volcano plot showing differentially expressed genes with p-value < 0.05 and fold change ≥ 2, comparing the control group to the HTPBDTRAP1 treatment group (b) and the control group to the HTPBDTRAP1 + 5-FU treatment group (c). d,e, Stacked plots of KEGG pathway analysis comparing the control group to the HTPBDTRAP1 treatment group (d) and the control group to the TPBDTRAP1 + 5-FU treatment group (e). Statistical significance was determined by unpaired two-tailed Student’s t-test (b).
Extended Data Fig. 3 Immune system pathways activated by HTPBDTRAP1 + 5-FU treatment.
a, Heat map for identification of differentially expressed genes in CRC tissues after control, HTPBDTRAP1 and HTPBDTRAP1 + 5-FU treatment. Three biological replicates are shown for each group. b,c, Bubble plots showing the major pathways associated with differentially expressed genes between the control and HTPBDTRAP1 + 5-FU treatment groups, as determined by KEGG enrichment analysis (b) and GO enrichment analysis (c). d,e, Chord plot (d) showing the KEGG enrichment analysis of differentially expressed genes between the control and HTPBDTRAP1 + 5-FU treatment groups, and representative pathway diagram (e) of GSEA enrichment analysis. Statistical significance was determined by Fisher’s exact test with Benjamini-Hochberg adjustment for multiple comparisons (b,c) and permutation test (e).
Extended Data Fig. 4 HTPBDTRAP1 + 5-FU treatment effectively modulates the tumour immune microenvironment.
a-e, Representative FCM plots and quantitative statistical analysis of DCs (CD80+CD86+ in CD11c+) in mesenteric lymph nodes (a), CD8+ cytotoxic T cells (CD8+ in CD3+) (b), Tregs (CD4+Foxp3+ in CD3+) (c), MDSCs (CD11b+Gr-1+) (d) or CD4+ helper T cells (CD4+ in CD3+) (e) in CRC tumour tissues after different treatments. n = 4 independent samples. f,g, The levels of IFN-γ, TNF-α (f), IL-6 and IL-12p70 (g) in CRC tumour tissues of different treatment groups were determined by ELISA. n = 4 independent samples. Data are presented as mean ± s.d. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc corrections (a-g).
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1–50, Tables 1–5 and References.
Supplementary Data
Statistical source data of Supplementary Information.
Source data
Source Data Fig. 1
Unprocessed western blots and statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, K., Yan, Y., Jin, XK. et al. An orally administered gene editing nanoparticle boosts chemo-immunotherapy in colorectal cancer. Nat. Nanotechnol. 20, 935–946 (2025). https://doi.org/10.1038/s41565-025-01904-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41565-025-01904-5
This article is cited by
-
Turning cold tumors into hot tumors to ignite immunotherapy
Molecular Cancer (2025)
-
Therapeutic targeting of cell death-immune crosstalk in cancer to rewire the tumor immune microenvironment
Molecular Cancer (2025)
-
Cyclophilin D suppresses colorectal cancer progression through the activation of an autophagy-mediated apoptotic pathway
Scientific Reports (2025)