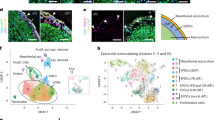
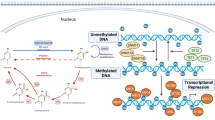
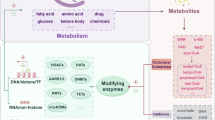
Abstract
Genetic and acquired forms of heart disease are leading causes of death worldwide. The epigenome, which governs cellular identity by modulating the accessibility of genetic regulatory elements, is established during development by transcription factors and has a pivotal role in the execution of cellular programmes. The epigenetic layers include DNA methylation, histone modifications and chromatin accessibility, which are dynamically regulated during development and in response to stress. Advances in single-cell and cell type-resolved epigenome analyses have provided unprecedented insights into the heterocellular nature of organs such as the heart, via the identification of epigenetic mechanisms and disease-associated epigenetic alterations in cardiomyocytes and other cardiac cell types. Chromatin remodelling, driven by specific modifiers, transcription factors and chaperones, orchestrates cardiac gene expression and contributes to disease manifestation and progression. Understanding how to modulate these epigenetic pathways in a cell type-specific manner offers promising avenues for therapeutic intervention, including epigenome editing for targeted modulation of regulatory elements. In this Review, we highlight studies decoding the various layers of the cardiac epigenome, emphasizing the interplay between cell type-specific mechanisms, describe emerging methods to study the cardiac epigenome, and discuss the translational potential of targeting epigenetic mechanisms for the prevention and treatment of cardiac diseases.
Key points
-
Epigenetic mechanisms govern transcriptional programmes in cardiac development and disease.
-
Different epigenetic layers control the establishment and activity of cardiac regulatory elements.
-
Cell type-specific or single-cell epigenome analysis is essential to decode mechanisms of epigenetic regulation in the heterocellular heart.
-
Epigenetic annotation provides insights into the disease pathogenesis underlying certain non-coding genetic variants.
-
Functional epigenetic editing and inhibition of epigenetic modulators provide novel avenues for treatment of heart disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
ENCODE Project Consortium. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Klose, R. J. & Bird, A. P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Bird, A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499–1504 (1980).
Ponger, L., Duret, L. & Mouchiroud, D. Determinants of CpG islands: expression in early embryo and isochore structure. Genome Res. 11, 1854–1860 (2001).
Angeloni, A. & Bogdanovic, O. Sequence determinants, function, and evolution of CpG islands. Biochem. Soc. Trans. 49, 1109–1119 (2021).
Isbel, L., Grand, R. S. & Schubeler, D. Generating specificity in genome regulation through transcription factor sensitivity to chromatin. Nat. Rev. Genet. 23, 728–740 (2022).
Domcke, S. et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579 (2015).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Movassagh, M. et al. Distinct epigenomic features in end-stage failing human hearts. Circulation 124, 2411–2422 (2011).
Movassagh, M. et al. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 5, e8564 (2010).
Kranzhofer, D. K. et al. 5′-Hydroxymethylcytosine precedes loss of CpG methylation in enhancers and genes undergoing activation in cardiomyocyte maturation. PLoS ONE 11, e0166575 (2016).
Greco, C. M. et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat. Commun. 7, 12418 (2016).
Laird, P. W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Morrison, J. et al. Evaluation of whole-genome DNA methylation sequencing library preparation protocols. Epigenetics Chromatin 14, 28 (2021).
Pidsley, R. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016).
Hon, G. C. et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206 (2013).
He, Y. et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature 583, 752–759 (2020).
Gilsbach, R. et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 9, 391 (2018).
Gunthel, M., Barnett, P. & Christoffels, V. M. Development, proliferation, and growth of the mammalian heart. Mol. Ther. 26, 1599–1609 (2018).
Ivey, M. J. et al. Resident fibroblast expansion during cardiac growth and remodeling. J. Mol. Cell Cardiol. 114, 161–174 (2018).
Meder, B. et al. Epigenome-Wide Association Study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation 136, 1528–1544 (2017).
Haas, J. et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 5, 413–429 (2013).
Pepin, M. E. et al. Racial and socioeconomic disparity associates with differences in cardiac DNA methylation among men with end-stage heart failure. Am. J. Physiol. Heart Circ. Physiol. 320, H2066–H2079 (2021).
Pepin, M. E. et al. DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure. Am. J. Physiol. Heart Circ. Physiol. 317, H674–H684 (2019).
Chapski, D. J. et al. Early adaptive chromatin remodeling events precede pathologic phenotypes and are reinforced in the failing heart. J. Mol. Cell. Cardiol. 160, 73–86 (2021).
Chen, H. et al. DNA methylation indicates susceptibility to isoproterenol-induced cardiac pathology and is associated with chromatin states. Circ. Res. 118, 786–797 (2016).
Oeing, C. U. et al. Indirect epigenetic testing identifies a diagnostic signature of cardiomyocyte DNA methylation in heart failure. Basic Res. Cardiol. 118, 9 (2023).
Baubec, T., Ivanek, R., Lienert, F. & Schubeler, D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 153, 480–492 (2013).
Hara, M. et al. Disturbance of cardiac gene expression and cardiomyocyte structure predisposes Mecp2-null mice to arrhythmias. Sci. Rep. 5, 11204 (2015).
Mayer, S. C. et al. Adrenergic repression of the epigenetic reader MeCP2 facilitates cardiac adaptation in chronic heart failure. Circ. Res. 117, 622–633 (2015).
Bin Akhtar, G., Buist, M. & Rastegar, M. MeCP2 and transcriptional control of eukaryotic gene expression. Eur. J. Cell Biol. 101, 151237 (2022).
Pastor, W. A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012).
Preissl, S., Gaulton, K. J. & Ren, B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat. Rev. Genet. 24, 21–43 (2023).
He, B. et al. Tissue-specific 5-hydroxymethylcytosine landscape of the human genome. Nat. Commun. 12, 4249 (2021).
Bhattacharyya, S. et al. Accurate classification of cardiomyopathy diagnosis by chromatin accessibility. Circulation 146, 878–881 (2022).
Tyagi, M., Imam, N., Verma, K. & Patel, A. K. Chromatin remodelers: we are the drivers!!. Nucleus 7, 388–404 (2016).
Scherba, J. C. et al. BRG1 is a biomarker of hypertrophic cardiomyopathy in human heart specimens. Sci. Rep. 12, 7996 (2022).
Alexander, J. M. et al. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 142, 1418–1430 (2015).
Hang, C. T. et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62–67 (2010).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Millan-Zambrano, G., Burton, A., Bannister, A. J. & Schneider, R. Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580 (2022).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
Raisner, R. et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 24, 1722–1729 (2018).
Yao, T. P. et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 (1998).
Shikama, N. et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 22, 5175–5185 (2003).
Miyamoto, S. et al. Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation 113, 679–690 (2006).
Blow, M. J. et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42, 806–810 (2010).
May, D. et al. Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 44, 89–93 (2011).
Papait, R. et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl Acad. Sci. USA 110, 20164–20169 (2013).
Hohl, M. et al. HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 123, 1359–1370 (2013).
Tan, W. L. W. et al. Epigenomes of human hearts reveal new genetic variants relevant for cardiac disease and phenotype. Circ. Res. 127, 761–777 (2020).
Pei, J. et al. H3K27ac acetylome signatures reveal the epigenomic reorganization in remodeled non-failing human hearts. Clin. Epigenetics 12, 106 (2020).
Dickel, D. E. et al. Genome-wide compendium and functional assessment of in vivo heart enhancers. Nat. Commun. 7, 12923 (2016).
He, A. et al. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat. Commun. 5, 4907 (2014).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Nord, A. S. et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013).
Hawe, J. S. et al. Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function. Nat. Genet. 54, 18–29 (2022).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Michels, K. B. et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat. Methods 10, 949–955 (2013).
Teschendorff, A. E., Zhu, T., Breeze, C. E. & Beck, S. EPISCORE: cell type deconvolution of bulk tissue DNA methylomes from single-cell RNA-Seq data. Genome Biol. 21, 221 (2020).
Schmidt, M., Maie, T., Dahl, E., Costa, I. G. & Wagner, W. Deconvolution of cellular subsets in human tissue based on targeted DNA methylation analysis at individual CpG sites. BMC Biol. 18, 178 (2020).
Titus, A. J., Gallimore, R. M., Salas, L. A. & Christensen, B. C. Cell-type deconvolution from DNA methylation: a review of recent applications. Hum. Mol. Genet. 26, R216–R224 (2017).
van den Oord, E., Xie, L. Y., Tran, C. J., Zhao, M. & Aberg, K. A. A targeted solution for estimating the cell-type composition of bulk samples. BMC Bioinform. 22, 462 (2021).
Lother, A. et al. Diabetes changes gene expression but not DNA methylation in cardiac cells. J. Mol. Cell. Cardiol. 151, 74–87 (2021).
Gilsbach, R. et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 5, 5288 (2014).
Preissl, S. et al. Deciphering the epigenetic code of cardiac myocyte transcription. Circ. Res. 117, 413–423 (2015).
Jugdutt, B. I. Ventricular remodeling after infarction and the extracellular collagen matrix: when is enough enough? Circulation 108, 1395–1403 (2003).
Pinto, A. R. et al. Revisiting cardiac cellular composition. Circ. Res. 118, 400–409 (2016).
Voigt, P., Tee, W. W. & Reinberg, D. A double take on bivalent promoters. Genes Dev. 27, 1318–1338 (2013).
Macrae, T. A., Fothergill-Robinson, J. & Ramalho-Santos, M. Regulation, functions and transmission of bivalent chromatin during mammalian development. Nat. Rev. Mol. Cell Biol. 24, 6–26 (2022).
Toker, L. et al. Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol. Neurodegener. 16, 31 (2021).
Mancarci, B. O. et al. Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro https://doi.org/10.1523/ENEURO.0212-17.2017 (2017).
Murphy, K. B., Ye, Y., Tsalenchuk, M., Nott, A. & Marzi, S. J. CHAS infers cell type-specific signatures in bulk brain histone acetylation studies of neurological and psychiatric disorders. Cell Rep. Methods 5, 101032 (2025).
Toker, L., Nido, G. S. & Tzoulis, C. Not every estimate counts - evaluation of cell composition estimation approaches in brain bulk tissue data. Genome Med. 15, 41 (2023).
Gasperini, M., Tome, J. M. & Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 21, 292–310 (2020).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001 e5919 (2021).
Hocker, J. D. et al. Cardiac cell type-specific gene regulatory programs and disease risk association. Sci. Adv. 7, eabf1444 (2021).
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
Ziller, M. J. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013).
Baubec, T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015).
Nuhrenberg, T. G. et al. Cardiac myocyte de novo DNA methyltransferases 3a/3b are dispensable for cardiac function and remodeling after chronic pressure overload in mice. PLoS ONE 10, e0131019 (2015).
Nothjunge, S. et al. DNA methylation signatures follow preformed chromatin compartments in cardiac myocytes. Nat. Commun. 8, 1667 (2017).
Vujic, A. et al. Experimental heart failure modelled by the cardiomyocyte-specific loss of an epigenome modifier, DNMT3B. J. Mol. Cell Cardiol. 82, 174–183 (2015).
Lahm, H. et al. Congenital heart disease risk loci identified by genome-wide association study in European patients. J. Clin. Invest. 131, e141837 (2021).
Stenzig, J. et al. Pharmacological inhibition of DNA methylation attenuates pressure overload-induced cardiac hypertrophy in rats. J. Mol. Cell. Cardiol. 120, 53–63 (2018).
Madsen, A. et al. An important role for DNMT3A-mediated DNA methylation in cardiomyocyte metabolism and contractility. Circulation 142, 1562–1578 (2020).
Madsen, A. et al. Hypertrophic signaling compensates for contractile and metabolic consequences of DNA methyltransferase 3A loss in human cardiomyocytes. J. Mol. Cell. Cardiol. 154, 115–123 (2021).
Tatton-Brown, K. et al. The Tatton-Brown–Rahman syndrome: a clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 3, 46 (2018).
Tatton-Brown, K. et al. Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet. 100, 725–736 (2017).
Stadler, M. B. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011).
Burger, L., Gaidatzis, D., Schubeler, D. & Stadler, M. B. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 41, e155 (2013).
Sonmezer, C. et al. Molecular co-occupancy identifies transcription factor binding cooperativity in vivo. Mol. Cell 81, 255–267 (2021).
Luna-Zurita, L. et al. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164, 999–1014 (2016).
He, A., Kong, S. W., Ma, Q. & Pu, W. T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl Acad. Sci. USA 108, 5632–5637 (2011).
Akerberg, B. N. et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat. Commun. 10, 4907 (2019).
Ang, Y. S. et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 167, 1734–1749 e1722 (2016).
Jurado Acosta, A. et al. Phosphorylation of GATA4 at serine 105 is required for left ventricular remodelling process in angiotensin II-induced hypertension in rats. Basic Clin. Pharmacol. Toxicol. 127, 178–195 (2020).
Liang, Q. et al. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276, 30245–30253 (2001).
Zhou, P., He, A. & Pu, W. T. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr. Top. Dev. Biol. 100, 143–169 (2012).
Hon, G. C. et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297 (2014).
Lan, Y. et al. Stage-specific regulation of DNA methylation by TET enzymes during human cardiac differentiation. Cell Rep. 37, 110095 (2021).
Fang, S. et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 10, 4297 (2019).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588 (2017).
Dahlet, T. et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat. Commun. 11, 3153 (2020).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Wamstad, J. A. et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206–220 (2012).
Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010).
Fu, J. D. et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 1, 235–247 (2013).
VanDusen, N. J. et al. Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation. Nat. Commun. 12, 4442 (2021).
Xin, M., Olson, E. N. & Bassel-Duby, R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14, 529–541 (2013).
Nguyen, A. T. et al. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 25, 263–274 (2011).
Cattaneo, P. et al. DOT1L regulates chamber-specific transcriptional networks during cardiogenesis and mediates postnatal cell cycle withdrawal. Nat. Commun. 13, 7444 (2022).
Hesse, M. et al. Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle. Nat. Commun. 3, 1076 (2012).
Monroe, T. O. et al. YAP partially reprograms chromatin accessibility to directly induce adult cardiogenesis in vivo. Dev. Cell 48, 765–779 (2019).
Chen, Y. et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science 373, 1537–1540 (2021).
Garry, G. A. & Olson, E. N. Reprogramming of cardiac cell fate as a therapeutic strategy for ischemic heart disease. J. Mol. Cell. Cardiol. 179, 2–6 (2023).
Hashimoto, H. et al. Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell 25, 69–86 (2019).
Gunthel, M. et al. Epigenetic state changes underlie metabolic switch in mouse post-infarction border zone cardiomyocytes. J. Cardiovasc. Dev. Dis. 8, 134 (2021).
Lee, D. P. et al. Robust CTCF-based chromatin architecture underpins epigenetic changes in the heart failure stress-gene response. Circulation 139, 1937–1956 (2019).
Zhang, Y. et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 51, 1380–1388 (2019).
Montefiori, L. E. et al. A promoter interaction map for cardiovascular disease genetics. eLife 7, e35788 (2018).
Greenwald, W. W. et al. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat. Commun. 10, 1054 (2019).
Bertero, A. et al. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 10, 1538 (2019).
Haydar, S. et al. Linking chamber-specific spatial chromatin interactions to disease variants and gene programs in human cardiomyocytes. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-5039927/v1 (2024).
Man, J. C. K. et al. Variant intronic enhancer controls SCN10A-short expression and heart conduction. Circulation 144, 229–242 (2021).
Man, J. C. K. et al. Genetic dissection of a super enhancer controlling the Nppa-Nppb cluster in the heart. Circ. Res. 128, 115–129 (2021).
Anene-Nzelu, C. G. et al. Assigning distal genomic enhancers to cardiac disease-causing genes. Circulation 142, 910–912 (2020).
Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA enhancer browser — a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007).
van Duijvenboden, K. et al. Conserved NPPB+ border zone switches from MEF2- to AP-1-driven gene program. Circulation 140, 864–879 (2019).
Leblanc, F. J. A. et al. Atrial fibrillation variant-to-gene prioritization through cross-ancestry eQTL and single-nucleus multiomic analyses. iScience 27, 110660 (2024).
Kosicki, M. et al. VISTA Enhancer browser: an updated database of tissue-specific developmental enhancers. Nucleic Acids Res. https://doi.org/10.1093/nar/gkae940 (2024).
Xiao, F. et al. Functional dissection of human cardiac enhancers and noncoding de novo variants in congenital heart disease. Nat. Genet. 56, 420–430 (2024).
Wang, Z. et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 22, 1131–1139 (2016).
Thienpont, B. et al. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J. Clin. Invest. 127, 335–348 (2017).
Papait, R. et al. Histone methyltransferase G9a is required for cardiomyocyte homeostasis and hypertrophy. Circulation 136, 1233–1246 (2017).
Gillette, T. G. & Hill, J. A. Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ. Res. 116, 1245–1253 (2015).
Neumayr, C. et al. Differential cofactor dependencies define distinct types of human enhancers. Nature 606, 406–413 (2022).
Litvinukova, M. et al. Cells of the adult human heart. Nature 588, 466–472 (2020).
Reichart, D. et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science 377, eabo1984 (2022).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Domcke, S. et al. A human cell atlas of fetal chromatin accessibility. Science 370, eaba7612 (2020).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Chan, A. S. F. et al. Spatio-temporal dynamics of the fibrotic niche in cardiac repair. Preprint at bioRxiv https://doi.org/10.1101/2024.11.10.622609 (2024).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Gaulton, K. J., Preissl, S. & Ren, B. Interpreting non-coding disease-associated human variants using single-cell epigenomics. Nat. Rev. Genet. 24, 516–534 (2023).
Cuomo, A. S. E., Nathan, A., Raychaudhuri, S., MacArthur, D. G. & Powell, J. E. Single-cell genomics meets human genetics. Nat. Rev. Genet. 24, 535–549 (2023).
Sweat, M. E. et al. Tbx5 maintains atrial identity in post-natal cardiomyocytes by regulating an atrial-specific enhancer network. Nat. Cardiovasc. Res. 2, 881–898 (2023).
Steimle, J. D. et al. Decoding the PITX2-controlled genetic network in atrial fibrillation. JCI Insight 7, e158895 (2022).
Zaidi, S. et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013).
Richter, F. et al. Genomic analyses implicate noncoding de novo variants in congenital heart disease. Nat. Genet. 52, 769–777 (2020).
Ameen, M. et al. Integrative single-cell analysis of cardiogenesis identifies developmental trajectories and non-coding mutations in congenital heart disease. Cell 185, 4937–4953 (2022).
Wang, L. et al. Single-cell dual-omics reveals the transcriptomic and epigenomic diversity of cardiac non-myocytes. Cardiovasc. Res. 118, 1548–1563 (2022).
Alexanian, M. et al. Chromatin remodelling drives immune cell-fibroblast communication in heart failure. Nature https://doi.org/10.1038/s41586-024-08085-6 (2024).
Alexanian, M. et al. A transcriptional switch governs fibroblast activation in heart disease. Nature 595, 438–443 (2021).
Amrute, J. M. et al. Defining cardiac functional recovery in end-stage heart failure at single-cell resolution. Nat. Cardiovasc. Res. 2, 399–416 (2023).
Amrute, J. M. et al. Targeting immune-fibroblast cell communication in heart failure. Nature https://doi.org/10.1038/s41586-024-08008-5 (2024).
Su, Q. et al. Single-cell insights: pioneering an integrated atlas of chromatin accessibility and transcriptomic landscapes in diabetic cardiomyopathy. Cardiovasc. Diabetol. 23, 139 (2024).
Ren, L. et al. Recent advances in epigenetic anticancer therapeutics and future perspectives. Front. Genet. 13, 1085391 (2022).
Perner, F., Gadrey, J. Y., Armstrong, S. A. & Kuhn, M. W. M. Targeting the Menin-KMT2A interaction in leukemia: lessons learned and future directions. Int. J. Cancer https://doi.org/10.1002/ijc.35332 (2025).
Abend, A. & Kehat, I. Histone deacetylases as therapeutic targets–from cancer to cardiac disease. Pharmacol. Ther. 147, 55–62 (2015).
Haldar, S. M. & McKinsey, T. A. BET-ting on chromatin-based therapeutics for heart failure. J. Mol. Cell Cardiol. 74, 98–102 (2014).
Kee, H. J. et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation 113, 51–59 (2006).
Granger, A. et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 22, 3549–3560 (2008).
Travers, J. G. et al. HDAC inhibition reverses preexisting diastolic dysfunction and blocks covert extracellular matrix remodeling. Circulation 143, 1874–1890 (2021).
Ranjbarvaziri, S. et al. Targeting HDAC6 to treat heart failure with preserved ejection fraction in mice. Nat. Commun. 15, 1352 (2024).
Lu, J., Qian, S. & Sun, Z. Targeting histone deacetylase in cardiac diseases. Front. Physiol. 15, 1405569 (2024).
Chun, P. Therapeutic effects of histone deacetylase inhibitors on heart disease. Arch. Pharm. Res. 43, 1276–1296 (2020).
McKinsey, T. A. et al. Emerging epigenetic therapies of cardiac fibrosis and remodelling in heart failure: from basic mechanisms to early clinical development. Cardiovasc. Res. 118, 3482–3498 (2023).
Jebessa, Z. H. et al. The lipid droplet-associated protein ABHD5 protects the heart through proteolysis of HDAC4. Nat. Metab. 1, 1157–1167 (2019).
Lehmann, L. H. et al. A proteolytic fragment of histone deacetylase 4 protects the heart from failure by regulating the hexosamine biosynthetic pathway. Nat. Med. 24, 62–72 (2018).
Finke, D. et al. Histone deacetylase 4 deletion broadly affects cardiac epigenetic repression and regulates transcriptional susceptibility via H3K9 methylation. J. Mol. Cell. Cardiol. 162, 119–129 (2022).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Padmanabhan, A. et al. BRD4 (bromodomain-containing protein 4) interacts with GATA4 (GATA binding protein 4) to govern mitochondrial homeostasis in adult cardiomyocytes. Circulation 142, 2338–2355 (2020).
Anand, P. et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Schwalm, M. P. & Knapp, S. BET bromodomain inhibitors. Curr. Opin. Chem. Biol. 68, 102148 (2022).
Pervaiz, M., Mishra, P. & Gunther, S. Bromodomain drug discovery - the past, the present, and the future. Chem. Rec. 18, 1808–1817 (2018).
Stratton, M. S. et al. Dynamic chromatin targeting of BRD4 stimulates cardiac fibroblast activation. Circ. Res. 125, 662–677 (2019).
Alexanian, M. et al. Chromatin remodelling drives immune cell-fibroblast communication in heart failure. Nature 635, 434–443 (2024).
Stratton, M. S., Haldar, S. M. & McKinsey, T. A. BRD4 inhibition for the treatment of pathological organ fibrosis. F1000Res 6, F1000 (2017).
Hsu, A. et al. Targeting transcription in heart failure via CDK7/12/13 inhibition. Nat. Commun. 13, 4345 (2022).
Nunez, J. K. et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519 (2021).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Schoger, E., Zimmermann, W. H., Cyganek, L. & Zelarayan, L. C. Establishment of a second generation homozygous CRISPRa human induced pluripotent stem cell (hiPSC) line for enhanced levels of endogenous gene activation. Stem Cell Res. 56, 102518 (2021).
Schoger, E. et al. CRISPR-mediated activation of endogenous gene expression in the postnatal heart. Circ. Res. 126, 6–24 (2020).
Laurette, P. et al. In vivo silencing of regulatory elements using a single AAV-CRISPRi vector. Circ. Res. 134, 223–225 (2024).
Han, J. L., Heinson, Y. W., Chua, C. J., Liu, W. & Entcheva, E. CRISPRi gene modulation and all-optical electrophysiology in post-differentiated human iPSC-cardiomyocytes. Commun. Biol. 6, 1236 (2023).
Amrute, J. M. et al. Single cell variant to enhancer to gene map for coronary artery disease. Preprint at medRxiv https://doi.org/10.1101/2024.11.13.24317257 (2024).
Deng, L. et al. Atlas of cardiac endothelial cell enhancer elements linking the mineralocorticoid receptor to pathological gene expression. Sci. Adv. 10, eadj5101 (2024).
Thakore, P. I. et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun. 9, 1674 (2018).
Tycko, J. et al. Development of compact transcriptional effectors using high-throughput measurements in diverse contexts. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02442-6 (2024).
Vora, S. et al. Rational design of a compact CRISPR-Cas9 activator for AAV-mediated delivery. Preprint at bioRxiv https://doi.org/10.1101/298620 (2018).
Weinmann, J. et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 11, 5432 (2020).
Mazurek, R. et al. AAV delivery strategy with mechanical support for safe and efficacious cardiac gene transfer in swine. Nat. Commun. 15, 10450 (2024).
Lek, A. et al. Death after high-dose rAAV9 gene therapy in a patient with Duchenne’s muscular dystrophy. N. Engl. J. Med. 389, 1203–1210 (2023).
Engreitz, J. M. et al. Deciphering the impact of genomic variation on function. Nature 633, 47–57 (2024).
Matharu, N. et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2019).
O’Connell, T. D., Rodrigo, M. C. & Simpson, P. C. Isolation and culture of adult mouse cardiac myocytes. Methods Mol. Biol. 357, 271–296 (2007).
Ackers-Johnson, M. & Foo, R. S. Langendorff-free isolation and propagation of adult mouse cardiomyocytes. Methods Mol. Biol. 1940, 193–204 (2019).
Denisenko, E. et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 130 (2020).
Li, H. et al. Optimized Langendorff perfusion system for cardiomyocyte isolation in adult mouse heart. J. Cell. Mol. Med. 24, 14619–14625 (2020).
Liu, B. et al. Comparative study on isolation and mitochondrial function of adult mouse and rat cardiomyocytes. J. Mol. Cell. Cardiol. 136, 64–71 (2019).
Nicks, A. M. et al. Standardised method for cardiomyocyte isolation and purification from individual murine neonatal, infant, and adult hearts. J. Mol. Cell. Cardiol. 170, 47–59 (2022).
Larcher, V. et al. An autofluorescence-based method for the isolation of highly purified ventricular cardiomyocytes. Cardiovasc. Res. 114, 409–416 (2018).
Bergmann, O. et al. Dynamics of cell generation and turnover in the human heart. Cell 161, 1566–1575 (2015).
Bergmann, O. et al. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp. Cell Res. 317, 188–194 (2011).
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009).
Robinson, E. L. et al. MSK-mediated phosphorylation of histone H3 Ser28 couples MAPK signalling with early gene induction and cardiac hypertrophy. Cells 11, 604 (2022).
Hill, M. C. & Martin, J. F. Epigenetic assays in purified cardiomyocyte nuclei. Methods Mol. Biol. 2158, 307–321 (2021).
See, K. et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat. Commun. 8, 225 (2017).
Cui, M. & Olson, E. N. Protocol for single-nucleus transcriptomics of diploid and tetraploid cardiomyocytes in murine hearts. STAR Protoc. 1, 100049 (2020).
Chatterjee, A. et al. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell 167, 722–738 (2016).
Cheedipudi, S. M. et al. Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ. Res. 124, 1198–1213 (2019).
Raulf, A. et al. Transgenic systems for unequivocal identification of cardiac myocyte nuclei and analysis of cardiomyocyte cell cycle status. Basic Res. Cardiol. 110, 33 (2015).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Wu, A. Z. et al. Phospholamban is concentrated in the nuclear envelope of cardiomyocytes and involved in perinuclear/nuclear calcium handling. J. Mol. Cell. Cardiol. 100, 1–8 (2016).
Hesse, M. et al. Proximity to injury, but neither number of nuclei nor ploidy define pathological adaptation and plasticity in cardiomyocytes. J. Mol. Cell. Cardiol. 152, 95–104 (2021).
Krane, M. et al. Sequential defects in cardiac lineage commitment and maturation cause hypoplastic left heart syndrome. Circulation 144, 1409–1428 (2021).
van Ouwerkerk, A. F. et al. Patient-specific TBX5-G125R variant induces profound transcriptional deregulation and atrial dysfunction. Circulation 145, 606–619 (2022).
Wirth, L. et al. Gene expression networks in endothelial cells from failing human hearts. Am. J. Physiol. Heart Circ. Physiol. 327, H573–H581 (2024).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Bhattacharyya, S., Sathe, A. A., Bhakta, M., Xing, C. & Munshi, N. V. PAN-INTACT enables direct isolation of lineage-specific nuclei from fibrous tissues. PLoS ONE 14, e0214677 (2019).
Yucel, N. et al. Cardiac endothelial cells maintain open chromatin and expression of cardiomyocyte myofibrillar genes. eLife 9, e55730 (2020).
Zhang, M., Lui, K. O. & Zhou, B. Application of new lineage tracing techniques in cardiovascular development and physiology. Circ. Res. 134, 445–458 (2024).
Yoshida, Y. & Yamanaka, S. Induced pluripotent stem cells 10 years later: for cardiac applications. Circ. Res. 120, 1958–1968 (2017).
Buikema, J. W. & Wu, S. M. Untangling the biology of genetic cardiomyopathies with pluripotent stem cell disease models. Curr. Cardiol. Rep. 19, 30 (2017).
Brandao, K. O., Tabel, V. A., Atsma, D. E., Mummery, C. L. & Davis, R. P. Human pluripotent stem cell models of cardiac disease: from mechanisms to therapies. Dis. Model. Mech. 10, 1039–1059 (2017).
Morton, S. U., Quiat, D., Seidman, J. G. & Seidman, C. E. Genomic frontiers in congenital heart disease. Nat. Rev. Cardiol. 19, 26–42 (2022).
Mills, R. J. & Hudson, J. E. Bioengineering adult human heart tissue: how close are we? APL Bioeng. 3, 010901 (2019).
Cho, S., Discher, D. E., Leong, K. W., Vunjak-Novakovic, G. & Wu, J. C. Challenges and opportunities for the next generation of cardiovascular tissue engineering. Nat. Methods https://doi.org/10.1038/s41592-022-01591-3 (2022).
Acknowledgements
The authors apologize to the authors whose essential contributions to the field of cardiac epigenetics could not be cited owing to space limitations. The authors received support from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) — SFB1550 — Project ID 464424253, A02: Collaborative Research Center 1550 (CRC1550) ‘Molecular Circuits of Heart Disease’, the SFB1425 — Project ID 422681845, P02/S03: Collaborative Research Center 1425 (CRC1425) ‘ScarCare’ and Project ID 558598989. They thank S. Preissl (University of Freiburg, Germany, and University of Graz, Austria) for discussing and proofreading the manuscript for initial submission, and J. Backs (University of Heidelberg, Germany) for supporting and discussing the initial concept of the manuscript.
Author information
Authors and Affiliations
Contributions
The authors contributed substantially to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cardiology thanks Vincent Christoffels and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- 5-Hydroxymethylcytosine
-
(5hmC). The first oxidized derivative of 5-methylcytosine (5mC) produced by ten-eleven translocation (TET) family enzymes during DNA demethylation in mammalian cells and associated with the establishment of accessible chromatin or regulatory activity.
- Accessible chromatin
-
Nucleosome-free genomic loci accessible to DNA-binding factors.
- Acquired heart disease
-
Heart disease that arises from non-genetic risk factors, such as metabolic syndrome, stress and hypertension.
- Chromatin
-
Complex of DNA and proteins that packages genomic information in the nucleus and regulates access to DNA-binding factors.
- Chromatin remodelling
-
Dynamic repositioning, exchange or eviction of nucleosomes to modulate DNA accessibility to transcription factors and other regulatory proteins.
- Cis-regulatory elements
-
(CREs). Non-coding DNA sequences bound by transcription factors and required for proper spatiotemporal regulation of target gene promoters in cis.
- Congenital heart disease
-
Structural and functional heart defects present at birth and caused by genetic and/or non-genetic causes.
- CpG
-
A dinucleotide consisting of a cytosine followed by a guanine. Cytosines in a CpG context are commonly methylated in mammalian cells. CpG methylation is inversely correlated with the regulatory activity of DNA-binding factors.
- DNA methylation
-
Methylation of DNA, most commonly at cytosine position 5 (5mC), predominantly at CpG dinucleotides in mammals and generally associated with transcriptional repression.
- Enhancers
-
Distal cis-regulatory elements that promote transcriptional activity of target genes, typically marked by histone 3 lysine 4 monomethylation (H3K4me1), H3K27 acetylation and low levels of CpG methylation.
- Epigenetic writers, erasers and readers
-
Proteins that can establish (writers), remove (erasers) or interpret (readers) epigenetic modifications, thereby regulating genome function and transcriptional activity.
- Epigenetics
-
Covers all inheritable changes in gene expression or phenotype that are not mediated by changes in the DNA sequence. The epigenetic layers in mammalian cells include DNA methylation, histone modifications and chromatin accessibility.
- Epigenome editing
-
Site-specific modification of the epigenome, mostly achieved by targeted recruitment of repressor or activator domains using CRISPR-based methods (CRISPR interference and CRISPR activation).
- Genetic heart disease
-
Heart disease elicited by inherited pathogenic genetic risk variants.
- Histone modification
-
Post-translational modification of histones is essential for epigenetic annotation of regulatory elements and is implicated in epigenetic regulation of gene expression.
- Induced pluripotent stem cell
-
(iPS cell). Somatic cell reprogrammed to a pluripotent state, which can be differentiated into different cell types, including cardiac cell types.
- Pioneering transcription factors
-
Transcription factors that can bind to inaccessible chromatin and initiate the establishment of regulatory elements during cell lineage specification.
- Promoters
-
Proximal regulatory elements adjacent to transcriptional start sites, typically marked by histone 3 lysine 4 trimethylation (H3K4me3), low levels of CpG methylation and accessible chromatin.
- Quantitative trait loci
-
Region of the genome associated with variability in quantitative traits, such as phenotype, gene expression, chromatin accessibility or histone acetylation.
- Silencers
-
Cis-regulatory elements that repress gene expression by recruiting transcriptional repressors and chromatin modifying and remodelling complexes.
- Transcription factors
-
DNA-binding proteins that modulate transcriptional activity.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laurette, P., Gilsbach, R. Cardiac epigenome in heart development and disease. Nat Rev Cardiol (2026). https://doi.org/10.1038/s41569-025-01223-1
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41569-025-01223-1